Johanna Ospel
A voxel-level approach to brain age prediction: A method to assess regional brain aging
Oct 17, 2023



Abstract:Brain aging is a regional phenomenon, a facet that remains relatively under-explored within the realm of brain age prediction research using machine learning methods. Voxel-level predictions can provide localized brain age estimates that can provide granular insights into the regional aging processes. This is essential to understand the differences in aging trajectories in healthy versus diseased subjects. In this work, a deep learning-based multitask model is proposed for voxel-level brain age prediction from T1-weighted magnetic resonance images. The proposed model outperforms the models existing in the literature and yields valuable clinical insights when applied to both healthy and diseased populations. Regional analysis is performed on the voxel-level brain age predictions to understand aging trajectories of known anatomical regions in the brain and show that there exist disparities in regional aging trajectories of healthy subjects compared to ones with underlying neurological disorders such as Dementia and more specifically, Alzheimer's disease. Our code is available at https://github.com/nehagianchandani/Voxel-level-brain-age-prediction.
Improved Segmentation and Detection Sensitivity of Diffusion-Weighted Brain Infarct Lesions with Synthetically Enhanced Deep Learning
Dec 29, 2020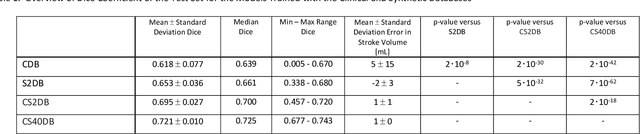
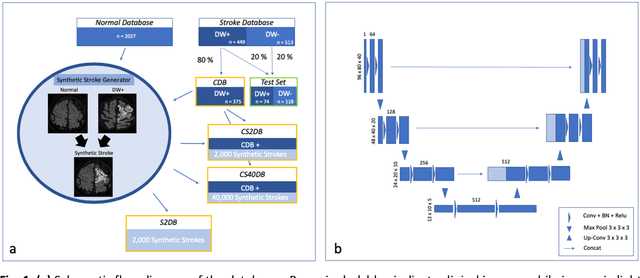
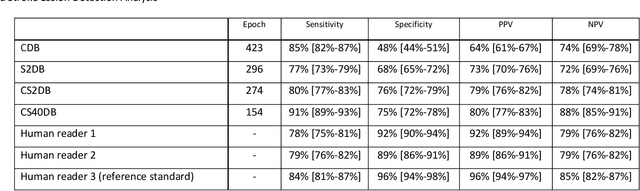
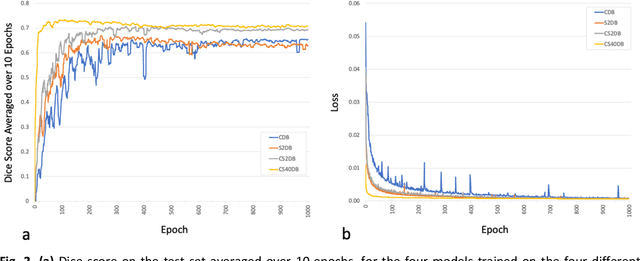
Abstract:Purpose: To compare the segmentation and detection performance of a deep learning model trained on a database of human-labelled clinical diffusion-weighted (DW) stroke lesions to a model trained on the same database enhanced with synthetic DW stroke lesions. Methods: In this institutional review board approved study, a stroke database of 962 cases (mean age 65+/-17 years, 255 males, 449 scans with DW positive stroke lesions) and a normal database of 2,027 patients (mean age 38+/-24 years,1088 females) were obtained. Brain volumes with synthetic DW stroke lesions were produced by warping the relative signal increase of real strokes to normal brain volumes. A generic 3D U-Net was trained on four different databases to generate four different models: (a) 375 neuroradiologist-labeled clinical DW positive stroke cases(CDB);(b) 2,000 synthetic cases(S2DB);(c) CDB+2,000 synthetic cases(CS2DB); or (d) CDB+40,000 synthetic cases(CS40DB). The models were tested on 20%(n=192) of the cases of the stroke database, which were excluded from the training set. Segmentation accuracy was characterized using Dice score and lesion volume of the stroke segmentation, and statistical significance was tested using a paired, two-tailed, Student's t-test. Detection sensitivity and specificity was compared to three neuroradiologists. Results: The performance of the 3D U-Net model trained on the CS40DB(mean Dice 0.72) was better than models trained on the CS2DB (0.70,P <0.001) or the CDB(0.65,P<0.001). The deep learning model was also more sensitive (91%[89%-93%]) than each of the three human readers(84%[81%-87%],78%[75%-81%],and 79%[76%-82%]), but less specific(75%[72%-78%] vs for the three human readers (96%[94%-97%],92%[90%-94%] and 89%[86%-91%]). Conclusion: Deep learning training for segmentation and detection of DW stroke lesions was significantly improved by enhancing the training set with synthetic lesions.
* This manuscript has been accepted for publication in Radiology: Artificial Intelligence
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge