Sebastian Kozerke
FlowMRI-Net: A generalizable self-supervised physics-driven 4D Flow MRI reconstruction network for aortic and cerebrovascular applications
Oct 11, 2024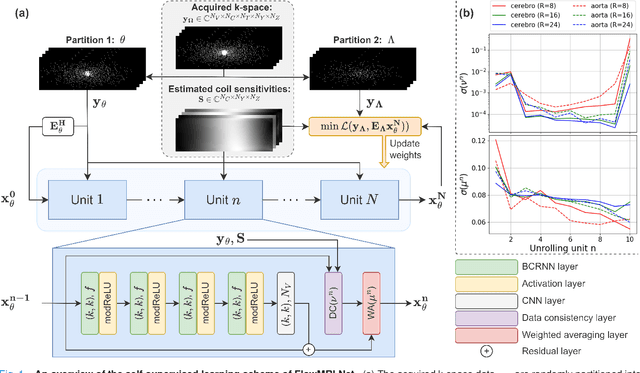
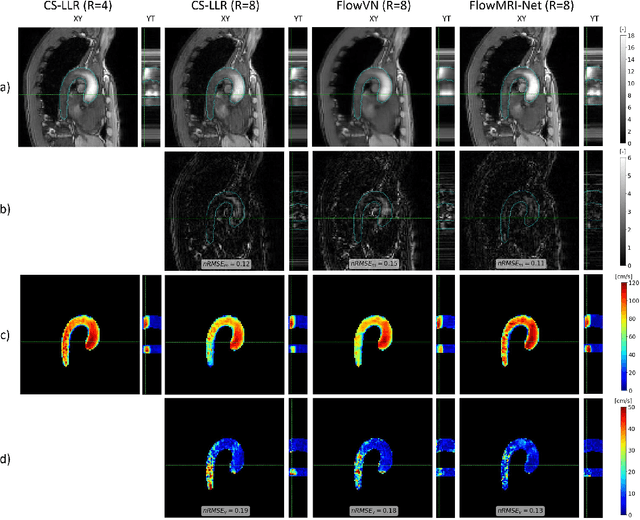
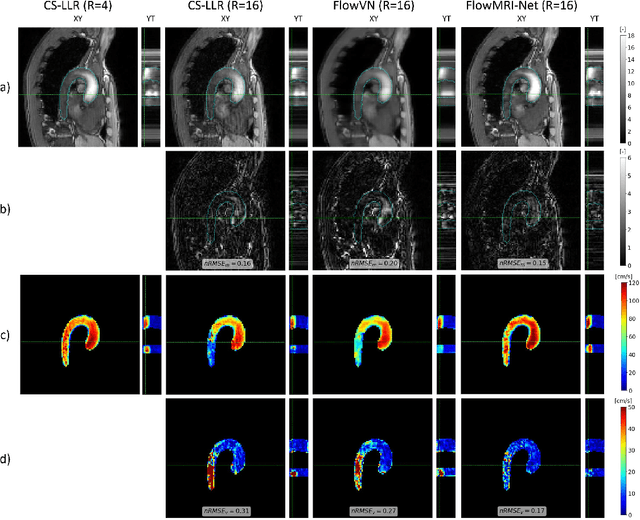
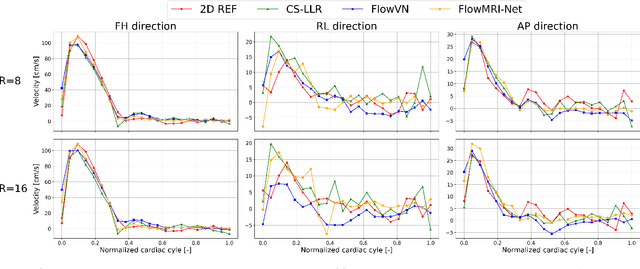
Abstract:In this work, we propose FlowMRI-Net, a novel deep learning-based framework for fast reconstruction of accelerated 4D flow magnetic resonance imaging (MRI) using physics-driven unrolled optimization and a complexvalued convolutional recurrent neural network trained in a self-supervised manner. The generalizability of the framework is evaluated using aortic and cerebrovascular 4D flow MRI acquisitions acquired on systems from two different vendors for various undersampling factors (R=8,16,24) and compared to state-of-the-art compressed sensing (CS-LLR) and deep learning-based (FlowVN) reconstructions. Evaluation includes quantitative analysis of image magnitudes, velocity magnitudes, and peak velocity curves. FlowMRINet outperforms CS-LLR and FlowVN for aortic 4D flow MRI reconstruction, resulting in vectorial normalized root mean square errors of $0.239\pm0.055$, $0.308\pm0.066$, and $0.302\pm0.085$ and mean directional errors of $0.023\pm0.015$, $0.036\pm0.018$, and $0.039\pm0.025$ for velocities in the thoracic aorta for R=16, respectively. Furthermore, FlowMRI-Net outperforms CS-LLR for cerebrovascular 4D flow MRI reconstruction, where no FlowVN can be trained due to the lack of a highquality reference, resulting in a consistent increase in SNR of around 6 dB and more accurate peak velocity curves for R=8,16,24. Reconstruction times ranged from 1 to 7 minutes on commodity CPU/GPU hardware. FlowMRI-Net enables fast and accurate quantification of aortic and cerebrovascular flow dynamics, with possible applications to other vascular territories. This will improve clinical adaptation of 4D flow MRI and hence may aid in the diagnosis and therapeutic management of cardiovascular diseases.
Optimal OnTheFly Feedback Control of Event Sensors
Aug 23, 2024
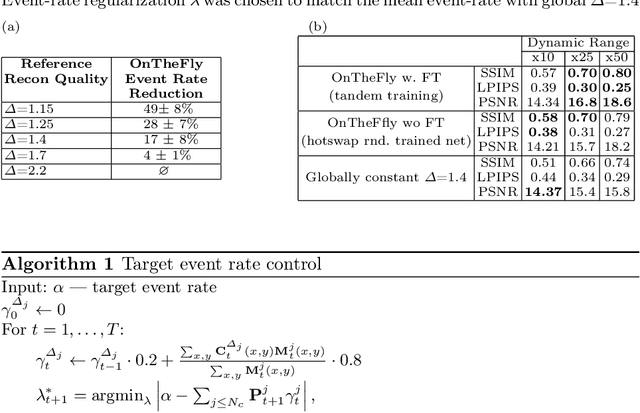
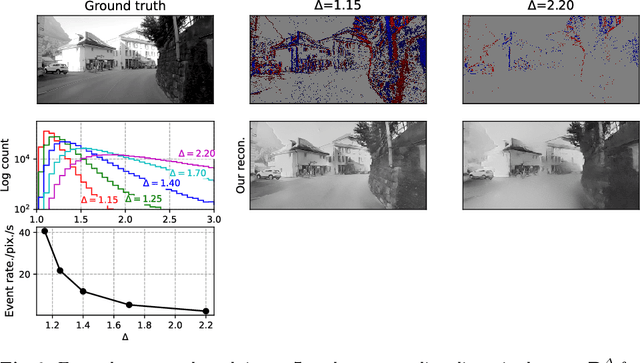
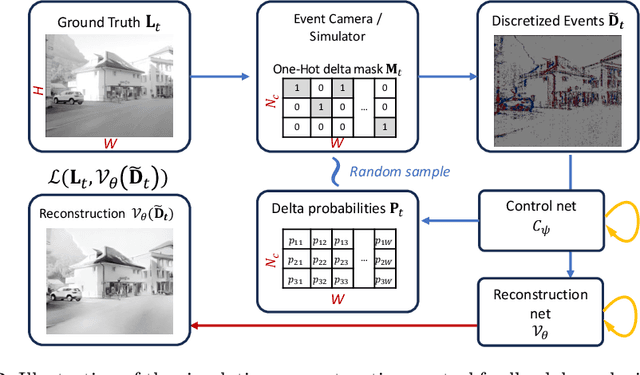
Abstract:Event-based vision sensors produce an asynchronous stream of events which are triggered when the pixel intensity variation exceeds a predefined threshold. Such sensors offer significant advantages, including reduced data redundancy, micro-second temporal resolution, and low power consumption, making them valuable for applications in robotics and computer vision. In this work, we consider the problem of video reconstruction from events, and propose an approach for dynamic feedback control of activation thresholds, in which a controller network analyzes the past emitted events and predicts the optimal distribution of activation thresholds for the following time segment. Additionally, we allow a user-defined target peak-event-rate for which the control network is conditioned and optimized to predict per-column activation thresholds that would eventually produce the best possible video reconstruction. The proposed OnTheFly control scheme is data-driven and trained in an end-to-end fashion using probabilistic relaxation of the discrete event representation. We demonstrate that our approach outperforms both fixed and randomly-varying threshold schemes by 6-12% in terms of LPIPS perceptual image dissimilarity metric, and by 49% in terms of event rate, achieving superior reconstruction quality while enabling a fine-tuned balance between performance accuracy and the event rate. Additionally, we show that sampling strategies provided by our OnTheFly control are interpretable and reflect the characteristics of the scene. Our results, derived from a physically-accurate simulator, underline the promise of the proposed methodology in enhancing the utility of event cameras for image reconstruction and other downstream tasks, paving the way for hardware implementation of dynamic feedback EVS control in silicon.
Improved Segmentation and Detection Sensitivity of Diffusion-Weighted Brain Infarct Lesions with Synthetically Enhanced Deep Learning
Dec 29, 2020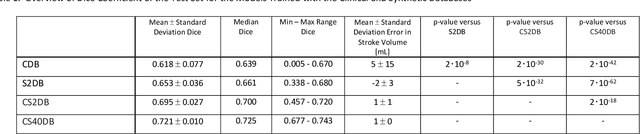
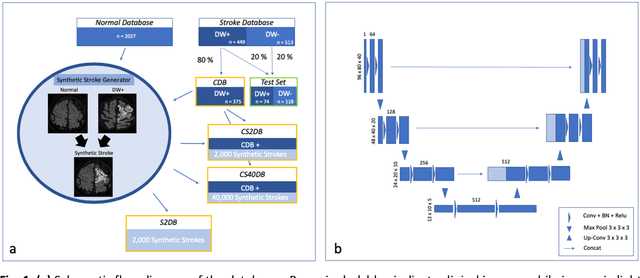
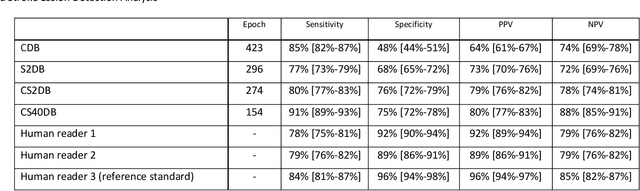
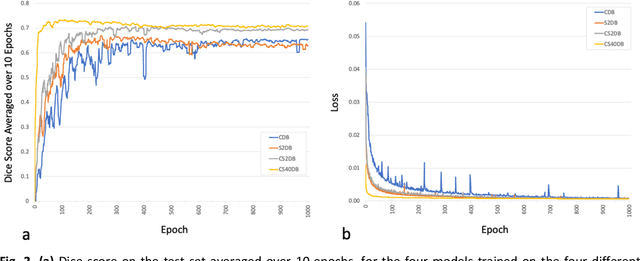
Abstract:Purpose: To compare the segmentation and detection performance of a deep learning model trained on a database of human-labelled clinical diffusion-weighted (DW) stroke lesions to a model trained on the same database enhanced with synthetic DW stroke lesions. Methods: In this institutional review board approved study, a stroke database of 962 cases (mean age 65+/-17 years, 255 males, 449 scans with DW positive stroke lesions) and a normal database of 2,027 patients (mean age 38+/-24 years,1088 females) were obtained. Brain volumes with synthetic DW stroke lesions were produced by warping the relative signal increase of real strokes to normal brain volumes. A generic 3D U-Net was trained on four different databases to generate four different models: (a) 375 neuroradiologist-labeled clinical DW positive stroke cases(CDB);(b) 2,000 synthetic cases(S2DB);(c) CDB+2,000 synthetic cases(CS2DB); or (d) CDB+40,000 synthetic cases(CS40DB). The models were tested on 20%(n=192) of the cases of the stroke database, which were excluded from the training set. Segmentation accuracy was characterized using Dice score and lesion volume of the stroke segmentation, and statistical significance was tested using a paired, two-tailed, Student's t-test. Detection sensitivity and specificity was compared to three neuroradiologists. Results: The performance of the 3D U-Net model trained on the CS40DB(mean Dice 0.72) was better than models trained on the CS2DB (0.70,P <0.001) or the CDB(0.65,P<0.001). The deep learning model was also more sensitive (91%[89%-93%]) than each of the three human readers(84%[81%-87%],78%[75%-81%],and 79%[76%-82%]), but less specific(75%[72%-78%] vs for the three human readers (96%[94%-97%],92%[90%-94%] and 89%[86%-91%]). Conclusion: Deep learning training for segmentation and detection of DW stroke lesions was significantly improved by enhancing the training set with synthetic lesions.
* This manuscript has been accepted for publication in Radiology: Artificial Intelligence
Deep variational network for rapid 4D flow MRI reconstruction
Apr 20, 2020
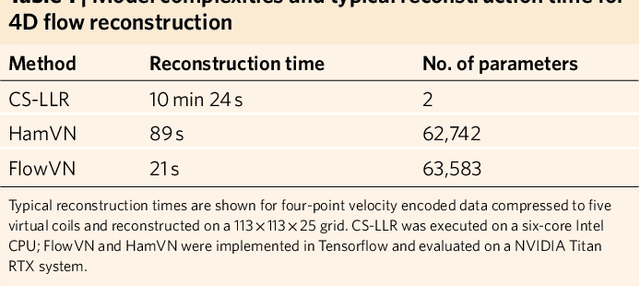
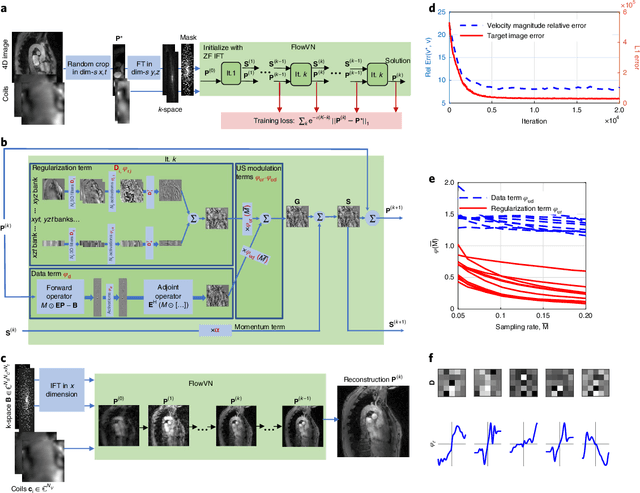

Abstract:Phase-contrast magnetic resonance imaging (MRI) provides time-resolved quantification of blood flow dynamics that can aid clinical diagnosis. Long in vivo scan times due to repeated three-dimensional (3D) volume sampling over cardiac phases and breathing cycles necessitate accelerated imaging techniques that leverage data correlations. Standard compressed sensing reconstruction methods require tuning of hyperparameters and are computationally expensive, which diminishes the potential reduction of examination times. We propose an efficient model-based deep neural reconstruction network and evaluate its performance on clinical aortic flow data. The network is shown to reconstruct undersampled 4D flow MRI data in under a minute on standard consumer hardware. Remarkably, the relatively low amounts of tunable parameters allowed the network to be trained on images from 11 reference scans while generalizing well to retrospective and prospective undersampled data for various acceleration factors and anatomies.
* 15 pages, 6 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge