Christian Federau
Random Expert Sampling for Deep Learning Segmentation of Acute Ischemic Stroke on Non-contrast CT
Sep 07, 2023Abstract:Purpose: Multi-expert deep learning training methods to automatically quantify ischemic brain tissue on Non-Contrast CT Materials and Methods: The data set consisted of 260 Non-Contrast CTs from 233 patients of acute ischemic stroke patients recruited in the DEFUSE 3 trial. A benchmark U-Net was trained on the reference annotations of three experienced neuroradiologists to segment ischemic brain tissue using majority vote and random expert sampling training schemes. We used a one-sided Wilcoxon signed-rank test on a set of segmentation metrics to compare bootstrapped point estimates of the training schemes with the inter-expert agreement and ratio of variance for consistency analysis. We further compare volumes with the 24h-follow-up DWI (final infarct core) in the patient subgroup with full reperfusion and we test volumes for correlation to the clinical outcome (mRS after 30 and 90 days) with the Spearman method. Results: Random expert sampling leads to a model that shows better agreement with experts than experts agree among themselves and better agreement than the agreement between experts and a majority-vote model performance (Surface Dice at Tolerance 5mm improvement of 61% to 0.70 +- 0.03 and Dice improvement of 25% to 0.50 +- 0.04). The model-based predicted volume similarly estimated the final infarct volume and correlated better to the clinical outcome than CT perfusion. Conclusion: A model trained on random expert sampling can identify the presence and location of acute ischemic brain tissue on Non-Contrast CT similar to CT perfusion and with better consistency than experts. This may further secure the selection of patients eligible for endovascular treatment in less specialized hospitals.
Latent Space Analysis of VAE and Intro-VAE applied to 3-dimensional MR Brain Volumes of Multiple Sclerosis, Leukoencephalopathy, and Healthy Patients
Jan 17, 2021
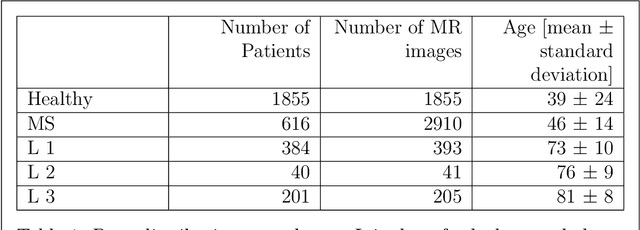


Abstract:Multiple Sclerosis (MS) and microvascular leukoencephalopathy are two distinct neurological conditions, the first caused by focal autoimmune inflammation in the central nervous system, the second caused by chronic white matter damage from atherosclerotic microvascular disease. Both conditions lead to signal anomalies on Fluid Attenuated Inversion Recovery (FLAIR) magnetic resonance (MR) images, which can be distinguished by an expert neuroradiologist, but which can look very similar to the untrained eye as well as in the early stage of both diseases. In this paper, we attempt to train a 3-dimensional deep neural network to learn the specific features of both diseases in an unsupervised manner. For this manner, in a first step we train a generative neural network to create artificial MR images of both conditions with approximate explicit density, using a mixed dataset of multiple sclerosis, leukoencephalopathy and healthy patients containing in total 5404 volumes of 3096 patients. In a second step, we distinguish features between the different diseases in the latent space of this network, and use them to classify new data.
Improved Segmentation and Detection Sensitivity of Diffusion-Weighted Brain Infarct Lesions with Synthetically Enhanced Deep Learning
Dec 29, 2020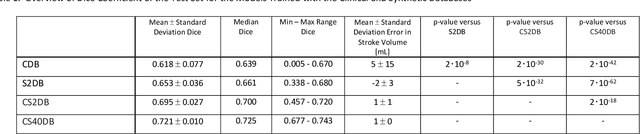
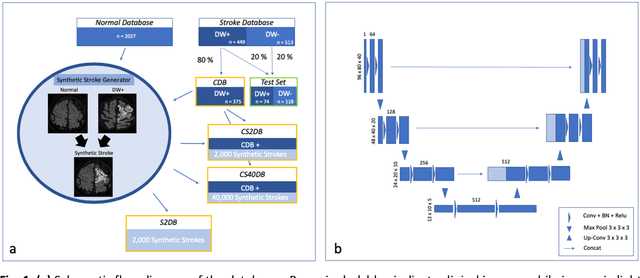
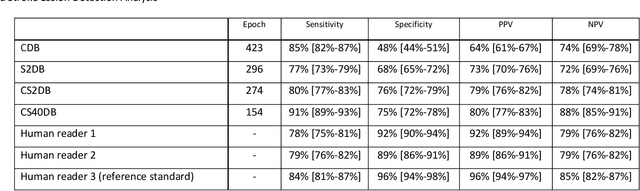
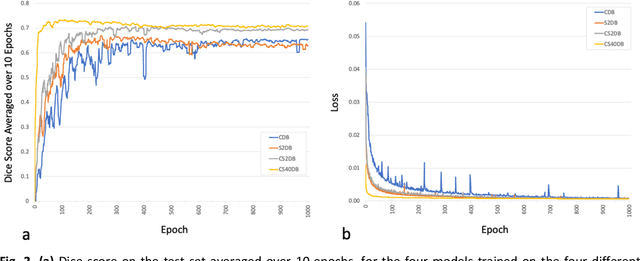
Abstract:Purpose: To compare the segmentation and detection performance of a deep learning model trained on a database of human-labelled clinical diffusion-weighted (DW) stroke lesions to a model trained on the same database enhanced with synthetic DW stroke lesions. Methods: In this institutional review board approved study, a stroke database of 962 cases (mean age 65+/-17 years, 255 males, 449 scans with DW positive stroke lesions) and a normal database of 2,027 patients (mean age 38+/-24 years,1088 females) were obtained. Brain volumes with synthetic DW stroke lesions were produced by warping the relative signal increase of real strokes to normal brain volumes. A generic 3D U-Net was trained on four different databases to generate four different models: (a) 375 neuroradiologist-labeled clinical DW positive stroke cases(CDB);(b) 2,000 synthetic cases(S2DB);(c) CDB+2,000 synthetic cases(CS2DB); or (d) CDB+40,000 synthetic cases(CS40DB). The models were tested on 20%(n=192) of the cases of the stroke database, which were excluded from the training set. Segmentation accuracy was characterized using Dice score and lesion volume of the stroke segmentation, and statistical significance was tested using a paired, two-tailed, Student's t-test. Detection sensitivity and specificity was compared to three neuroradiologists. Results: The performance of the 3D U-Net model trained on the CS40DB(mean Dice 0.72) was better than models trained on the CS2DB (0.70,P <0.001) or the CDB(0.65,P<0.001). The deep learning model was also more sensitive (91%[89%-93%]) than each of the three human readers(84%[81%-87%],78%[75%-81%],and 79%[76%-82%]), but less specific(75%[72%-78%] vs for the three human readers (96%[94%-97%],92%[90%-94%] and 89%[86%-91%]). Conclusion: Deep learning training for segmentation and detection of DW stroke lesions was significantly improved by enhancing the training set with synthetic lesions.
* This manuscript has been accepted for publication in Radiology: Artificial Intelligence
Image Translation for Medical Image Generation -- Ischemic Stroke Lesions
Oct 05, 2020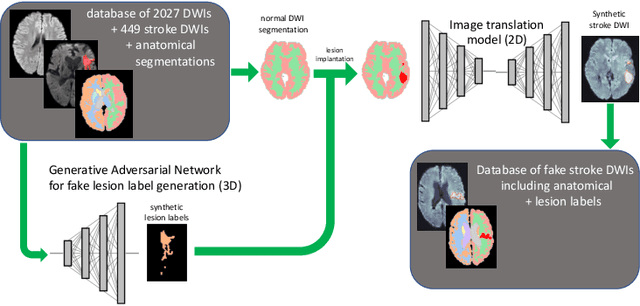
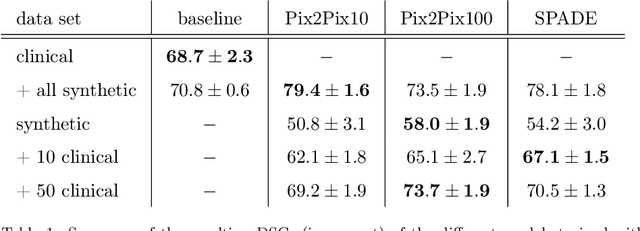
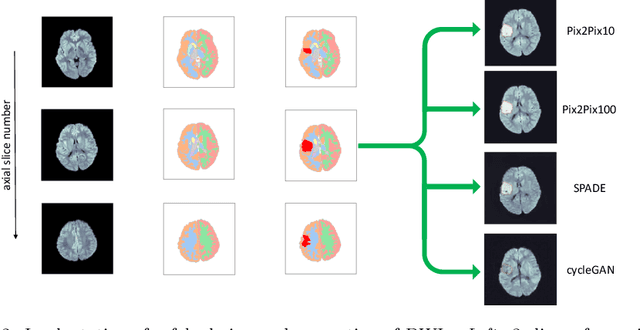
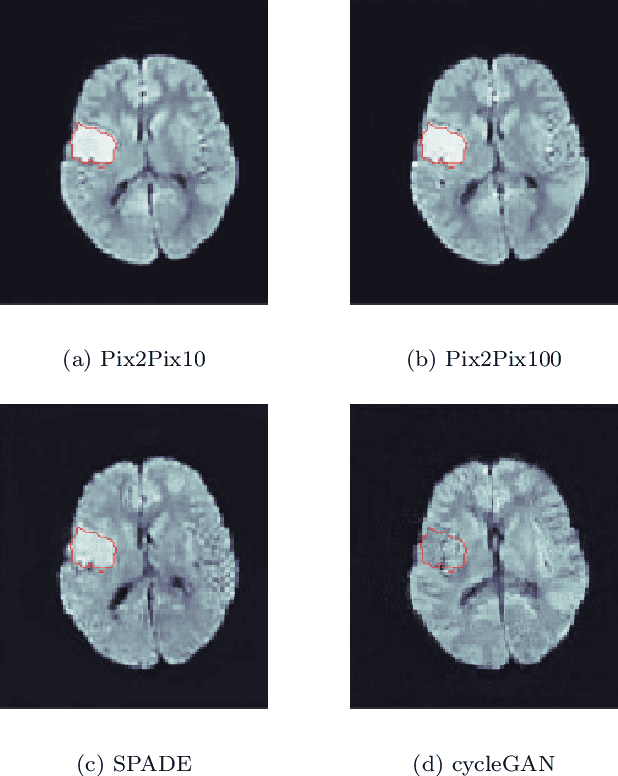
Abstract:Deep learning-based automated disease detection and segmentation algorithms promise to accelerate and improve many clinical processes. However, such algorithms require vast amounts of annotated training data, which are typically not available in a medical context, e.g., due to data privacy concerns, legal obstructions, and non-uniform data formats. Synthetic databases of annotated pathologies could provide the required amounts of training data. Here, we demonstrate with the example of ischemic stroke that a significant improvement in lesion segmentation is feasible using deep learning-based data augmentation. To this end, we train different image-to-image translation models to synthesize diffusion-weighted magnetic resonance images (DWIs) of brain volumes with and without stroke lesions from semantic segmentation maps. In addition, we train a generative adversarial network to generate synthetic lesion masks. Subsequently, we combine these two components to build a large database of synthetic stroke DWIs. The performance of the various generative models is evaluated using a U-Net which is trained to segment stroke lesions on a clinical test set. We compare the results to human expert inter-reader scores. For the model with the best performance, we report a maximum Dice score of 82.6\%, which significantly outperforms the model trained on the clinical images alone (74.8\%), and also the inter-reader Dice score of two human readers of 76.9\%. Moreover, we show that for a very limited database of only 10 or 50 clinical cases, synthetic data can be used to pre-train the segmentation algorithms, which ultimately yields an improvement by a factor of as high as 8 compared to a setting where no synthetic data is used.
Multi-modal segmentation of 3D brain scans using neural networks
Aug 11, 2020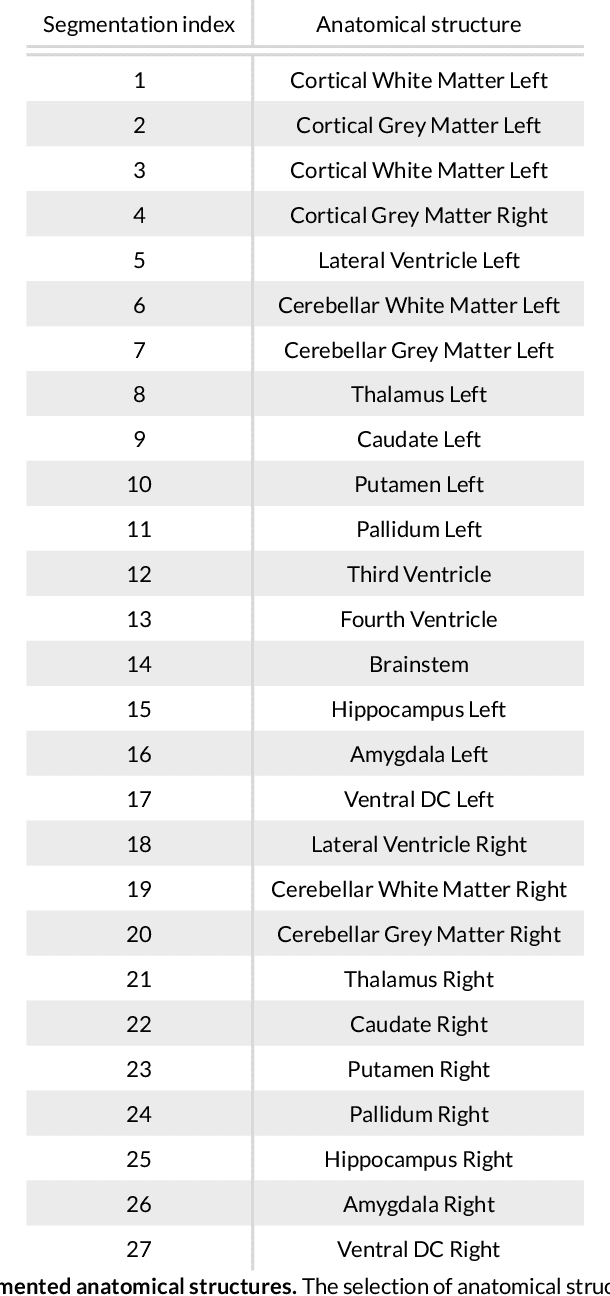
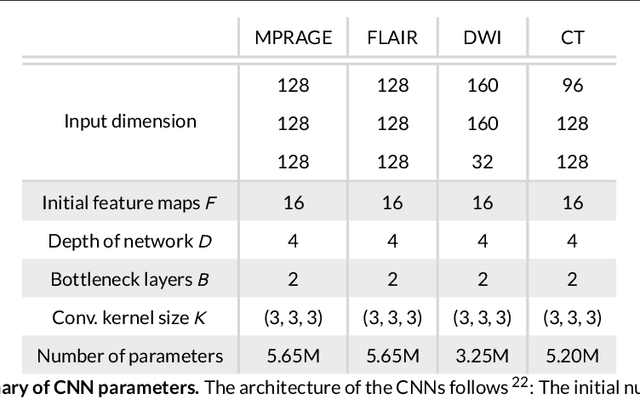
Abstract:Purpose: To implement a brain segmentation pipeline based on convolutional neural networks, which rapidly segments 3D volumes into 27 anatomical structures. To provide an extensive, comparative study of segmentation performance on various contrasts of magnetic resonance imaging (MRI) and computed tomography (CT) scans. Methods: Deep convolutional neural networks are trained to segment 3D MRI (MPRAGE, DWI, FLAIR) and CT scans. A large database of in total 851 MRI/CT scans is used for neural network training. Training labels are obtained on the MPRAGE contrast and coregistered to the other imaging modalities. The segmentation quality is quantified using the Dice metric for a total of 27 anatomical structures. Dropout sampling is implemented to identify corrupted input scans or low-quality segmentations. Full segmentation of 3D volumes with more than 2 million voxels is obtained in less than 1s of processing time on a graphical processing unit. Results: The best average Dice score is found on $T_1$-weighted MPRAGE ($85.3\pm4.6\,\%$). However, for FLAIR ($80.0\pm7.1\,\%$), DWI ($78.2\pm7.9\,\%$) and CT ($79.1\pm 7.9\,\%$), good-quality segmentation is feasible for most anatomical structures. Corrupted input volumes or low-quality segmentations can be detected using dropout sampling. Conclusion: The flexibility and performance of deep convolutional neural networks enables the direct, real-time segmentation of FLAIR, DWI and CT scans without requiring $T_1$-weighted scans.
Diffusion-Weighted Magnetic Resonance Brain Images Generation with Generative Adversarial Networks and Variational Autoencoders: A Comparison Study
Jun 24, 2020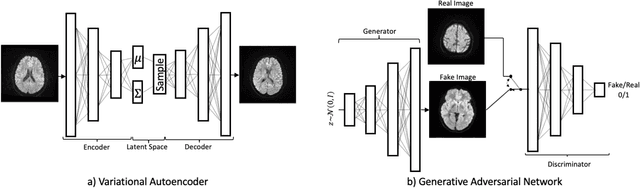
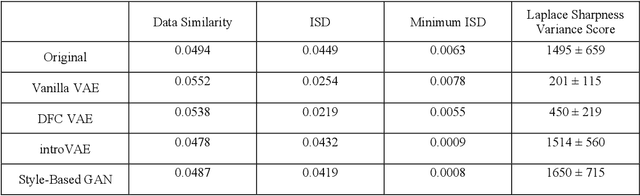
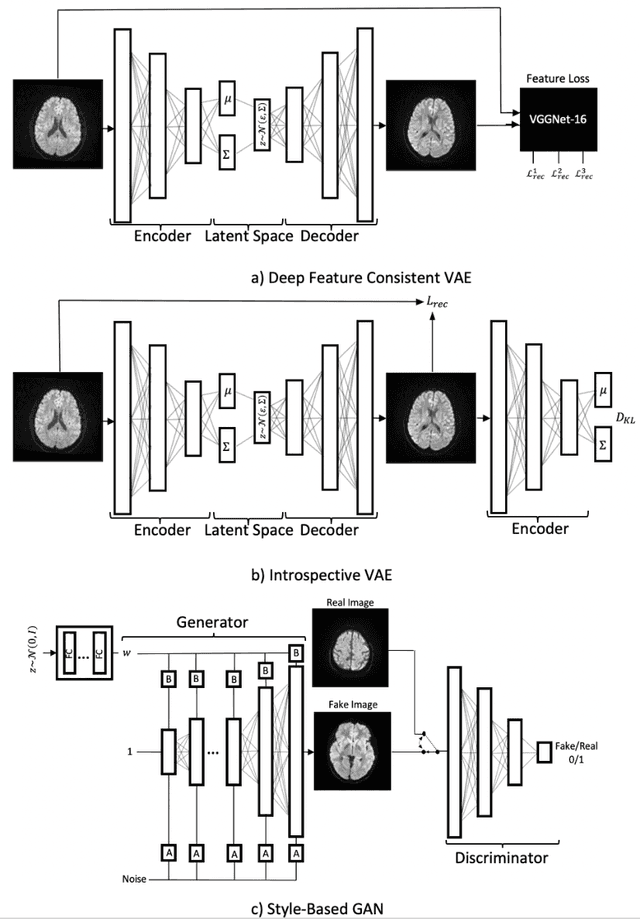
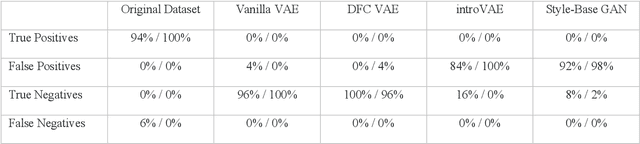
Abstract:We show that high quality, diverse and realistic-looking diffusion-weighted magnetic resonance images can be synthesized using deep generative models. Based on professional neuroradiologists' evaluations and diverse metrics with respect to quality and diversity of the generated synthetic brain images, we present two networks, the Introspective Variational Autoencoder and the Style-Based GAN, that qualify for data augmentation in the medical field, where information is saved in a dispatched and inhomogeneous way and access to it is in many aspects restricted.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge