Jonathan Zopes
Image Translation for Medical Image Generation -- Ischemic Stroke Lesions
Oct 05, 2020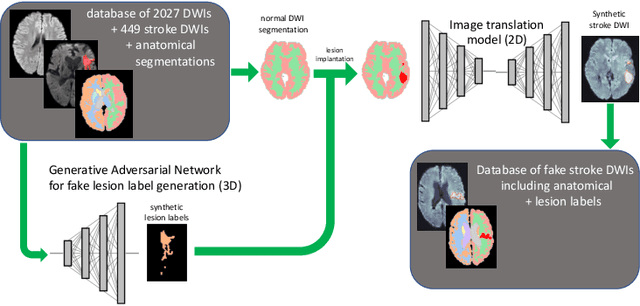
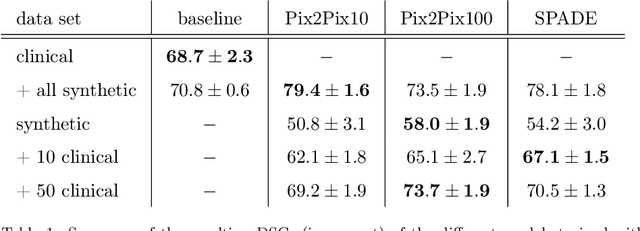
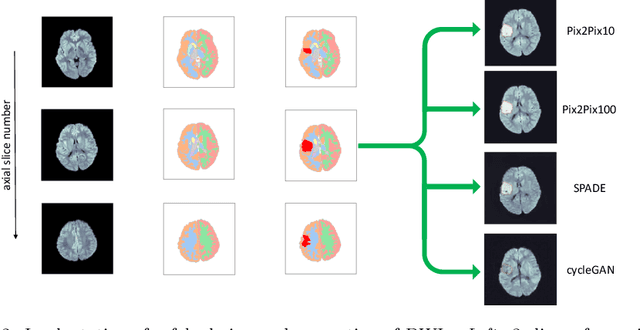
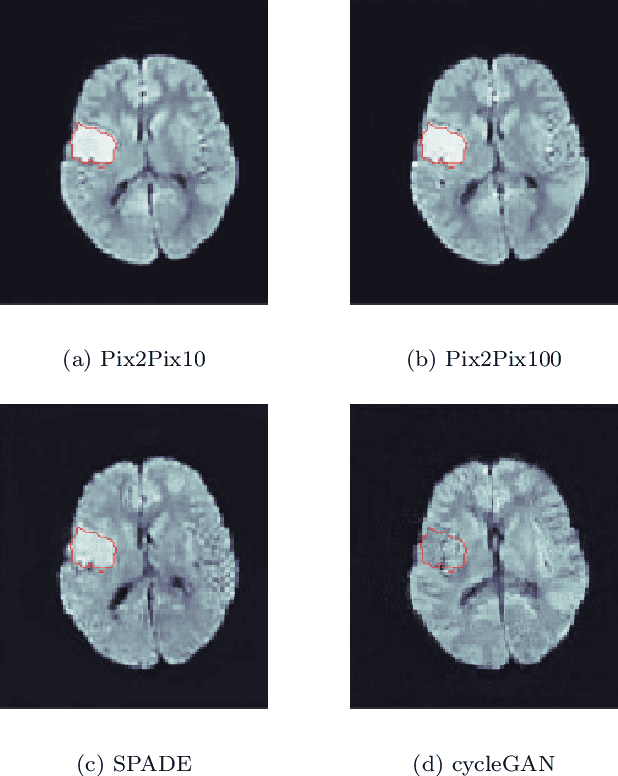
Abstract:Deep learning-based automated disease detection and segmentation algorithms promise to accelerate and improve many clinical processes. However, such algorithms require vast amounts of annotated training data, which are typically not available in a medical context, e.g., due to data privacy concerns, legal obstructions, and non-uniform data formats. Synthetic databases of annotated pathologies could provide the required amounts of training data. Here, we demonstrate with the example of ischemic stroke that a significant improvement in lesion segmentation is feasible using deep learning-based data augmentation. To this end, we train different image-to-image translation models to synthesize diffusion-weighted magnetic resonance images (DWIs) of brain volumes with and without stroke lesions from semantic segmentation maps. In addition, we train a generative adversarial network to generate synthetic lesion masks. Subsequently, we combine these two components to build a large database of synthetic stroke DWIs. The performance of the various generative models is evaluated using a U-Net which is trained to segment stroke lesions on a clinical test set. We compare the results to human expert inter-reader scores. For the model with the best performance, we report a maximum Dice score of 82.6\%, which significantly outperforms the model trained on the clinical images alone (74.8\%), and also the inter-reader Dice score of two human readers of 76.9\%. Moreover, we show that for a very limited database of only 10 or 50 clinical cases, synthetic data can be used to pre-train the segmentation algorithms, which ultimately yields an improvement by a factor of as high as 8 compared to a setting where no synthetic data is used.
Multi-modal segmentation of 3D brain scans using neural networks
Aug 11, 2020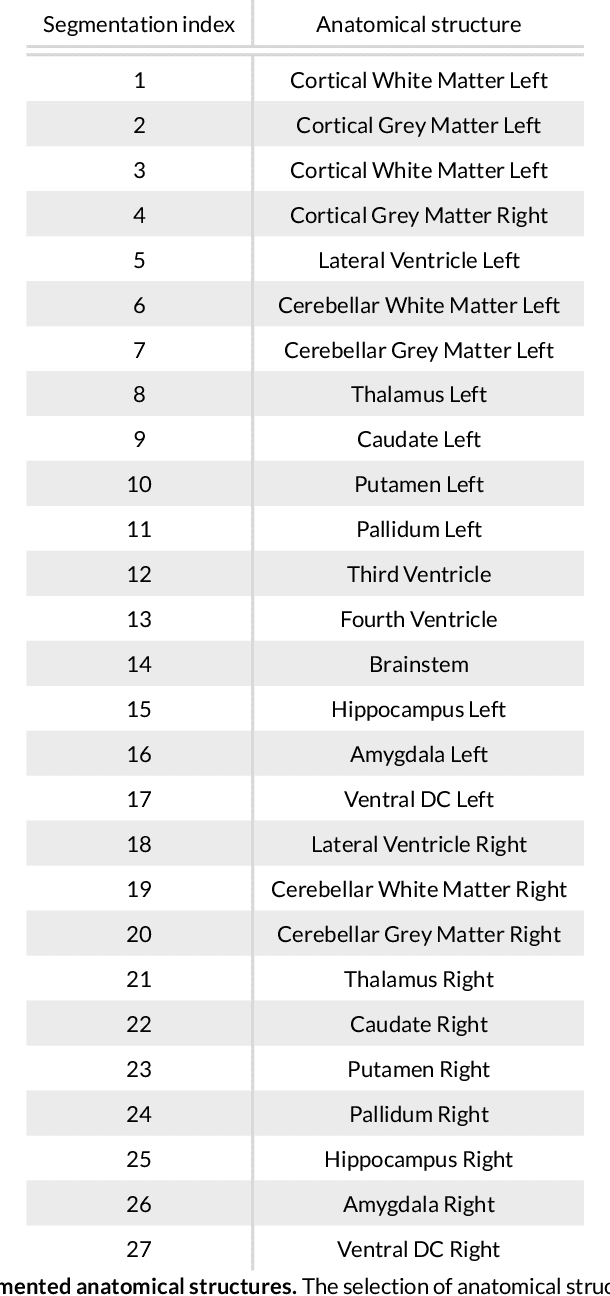
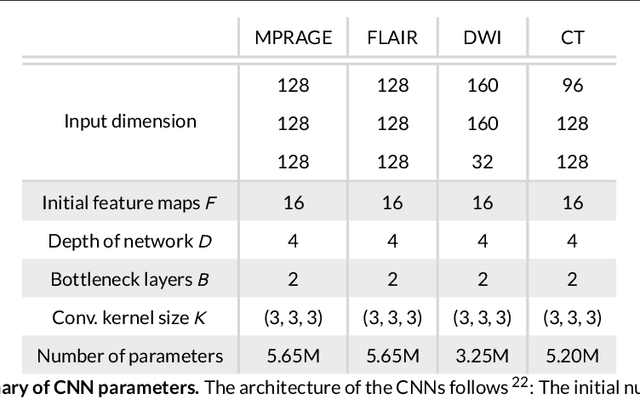
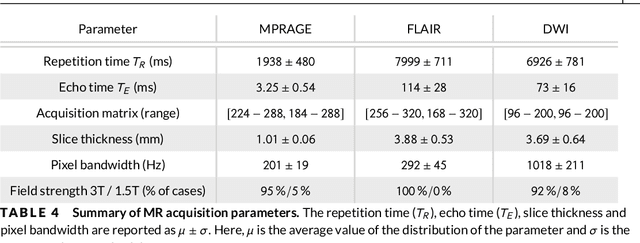
Abstract:Purpose: To implement a brain segmentation pipeline based on convolutional neural networks, which rapidly segments 3D volumes into 27 anatomical structures. To provide an extensive, comparative study of segmentation performance on various contrasts of magnetic resonance imaging (MRI) and computed tomography (CT) scans. Methods: Deep convolutional neural networks are trained to segment 3D MRI (MPRAGE, DWI, FLAIR) and CT scans. A large database of in total 851 MRI/CT scans is used for neural network training. Training labels are obtained on the MPRAGE contrast and coregistered to the other imaging modalities. The segmentation quality is quantified using the Dice metric for a total of 27 anatomical structures. Dropout sampling is implemented to identify corrupted input scans or low-quality segmentations. Full segmentation of 3D volumes with more than 2 million voxels is obtained in less than 1s of processing time on a graphical processing unit. Results: The best average Dice score is found on $T_1$-weighted MPRAGE ($85.3\pm4.6\,\%$). However, for FLAIR ($80.0\pm7.1\,\%$), DWI ($78.2\pm7.9\,\%$) and CT ($79.1\pm 7.9\,\%$), good-quality segmentation is feasible for most anatomical structures. Corrupted input volumes or low-quality segmentations can be detected using dropout sampling. Conclusion: The flexibility and performance of deep convolutional neural networks enables the direct, real-time segmentation of FLAIR, DWI and CT scans without requiring $T_1$-weighted scans.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge