Yongkai Liu
Random Expert Sampling for Deep Learning Segmentation of Acute Ischemic Stroke on Non-contrast CT
Sep 07, 2023Abstract:Purpose: Multi-expert deep learning training methods to automatically quantify ischemic brain tissue on Non-Contrast CT Materials and Methods: The data set consisted of 260 Non-Contrast CTs from 233 patients of acute ischemic stroke patients recruited in the DEFUSE 3 trial. A benchmark U-Net was trained on the reference annotations of three experienced neuroradiologists to segment ischemic brain tissue using majority vote and random expert sampling training schemes. We used a one-sided Wilcoxon signed-rank test on a set of segmentation metrics to compare bootstrapped point estimates of the training schemes with the inter-expert agreement and ratio of variance for consistency analysis. We further compare volumes with the 24h-follow-up DWI (final infarct core) in the patient subgroup with full reperfusion and we test volumes for correlation to the clinical outcome (mRS after 30 and 90 days) with the Spearman method. Results: Random expert sampling leads to a model that shows better agreement with experts than experts agree among themselves and better agreement than the agreement between experts and a majority-vote model performance (Surface Dice at Tolerance 5mm improvement of 61% to 0.70 +- 0.03 and Dice improvement of 25% to 0.50 +- 0.04). The model-based predicted volume similarly estimated the final infarct volume and correlated better to the clinical outcome than CT perfusion. Conclusion: A model trained on random expert sampling can identify the presence and location of acute ischemic brain tissue on Non-Contrast CT similar to CT perfusion and with better consistency than experts. This may further secure the selection of patients eligible for endovascular treatment in less specialized hospitals.
A Subabdominal MRI Image Segmentation Algorithm Based on Multi-Scale Feature Pyramid Network and Dual Attention Mechanism
May 19, 2023Abstract:This study aimed to solve the semantic gap and misalignment issue between encoding and decoding because of multiple convolutional and pooling operations in U-Net when segmenting subabdominal MRI images during rectal cancer treatment. A MRI Image Segmentation is proposed based on a multi-scale feature pyramid network and dual attention mechanism. Our innovation is the design of two modules: 1) a dilated convolution and multi-scale feature pyramid network are used in the encoding to avoid the semantic gap. 2) a dual attention mechanism is designed to maintain spatial information of U-Net and reduce misalignment. Experiments on a subabdominal MRI image dataset show the proposed method achieves better performance than others methods. In conclusion, a multi-scale feature pyramid network can reduce the semantic gap, and the dual attention mechanism can make an alignment of features between encoding and decoding.
Decentralized Coverage Path Planning with Reinforcement Learning and Dual Guidance
Oct 14, 2022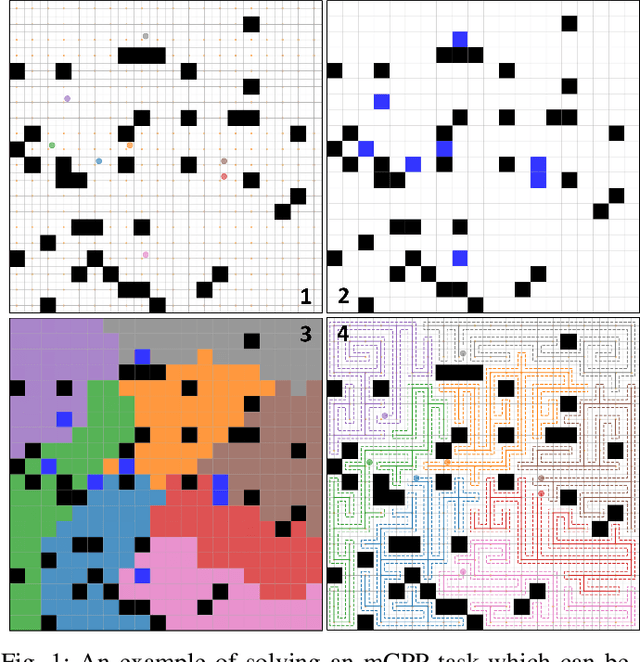
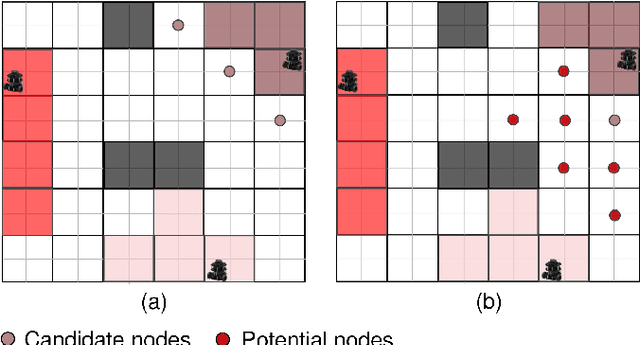
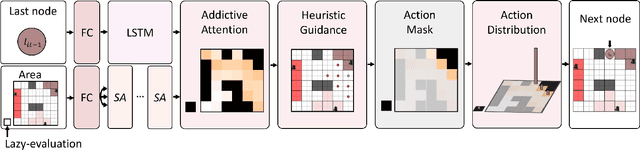
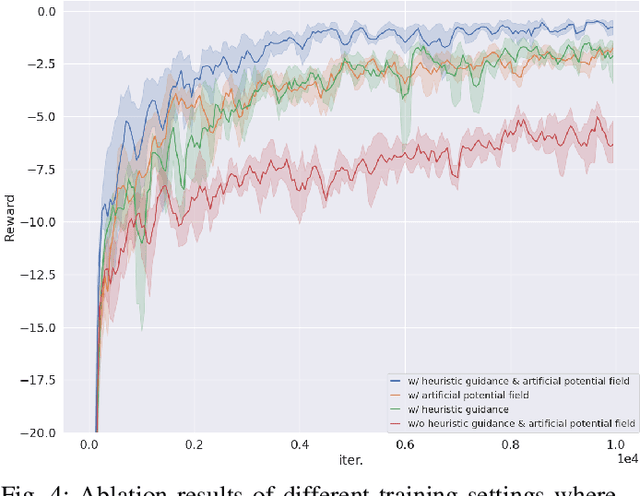
Abstract:Planning coverage path for multiple robots in a decentralized way enhances robustness to coverage tasks handling uncertain malfunctions. To achieve high efficiency in a distributed manner for each single robot, a comprehensive understanding of both the complicated environments and cooperative agents intent is crucial. Unfortunately, existing works commonly consider only part of these factors, resulting in imbalanced subareas or unnecessary overlaps. To tackle this issue, we introduce a Decentralized reinforcement learning framework with dual guidance to train each agent to solve the decentralized multiple coverage path planning problem straightly through the environment states. As distributed robots require others intentions to perform better coverage efficiency, we utilize two guidance methods, artificial potential fields and heuristic guidance, to include and integrate others intentions into observations for each robot. With our constructed framework, results have shown our agents successfully learn to determine their own subareas while achieving full coverage, balanced subareas and low overlap rates. We then implement spanning tree cover within those subareas to construct actual routes for each robot and complete given coverage tasks. Our performance is also compared with the state of the art decentralized method showing at most 10 percent lower overlap rates while performing high efficiency in similar environments.
ME-Net: Multi-Encoder Net Framework for Brain Tumor Segmentation
Mar 21, 2022Abstract:Glioma is the most common and aggressive brain tumor. Magnetic resonance imaging (MRI) plays a vital role to evaluate tumors for the arrangement of tumor surgery and the treatment of subsequent procedures. However, the manual segmentation of the MRI image is strenuous, which limits its clinical application. With the development of deep learning, a large number of automatic segmentation methods have been developed, but most of them stay in 2D images, which leads to subpar performance. Moreover, the serious voxel imbalance between the brain tumor and the background as well as the different sizes and locations of the brain tumor makes the segmentation of 3D images a challenging problem. Aiming at segmenting 3D MRI, we propose a model for brain tumor segmentation with multiple encoders. The structure contains four encoders and one decoder. The four encoders correspond to the four modalities of the MRI image, perform one-to-one feature extraction, and then merge the feature maps of the four modalities into the decoder. This method reduces the difficulty of feature extraction and greatly improves model performance. We also introduced a new loss function named "Categorical Dice", and set different weights for different segmented regions at the same time, which solved the problem of voxel imbalance. We evaluated our approach using the online BraTS 2020 Challenge verification. Our proposed method can achieve promising results in the validation set compared to the state-of-the-art approaches with Dice scores of 0.70249, 0.88267, and 0.73864 for the intact tumor, tumor core, and enhanced tumor, respectively.
Automatic Prostate Zonal Segmentation Using Fully Convolutional Network with Feature Pyramid Attention
Oct 31, 2019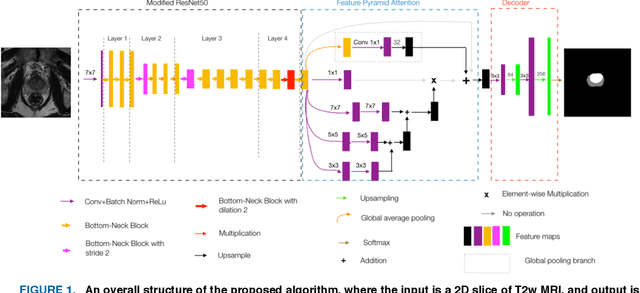
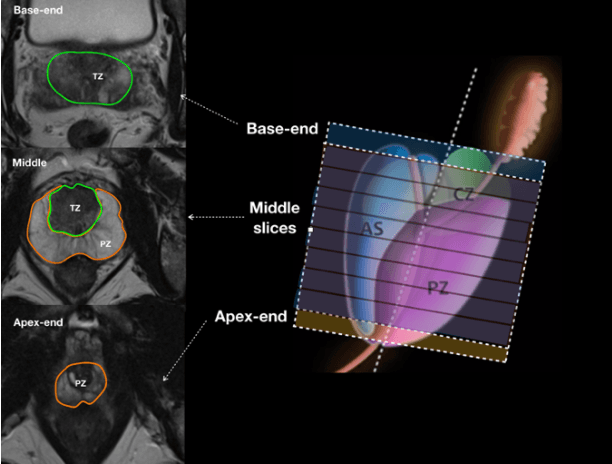
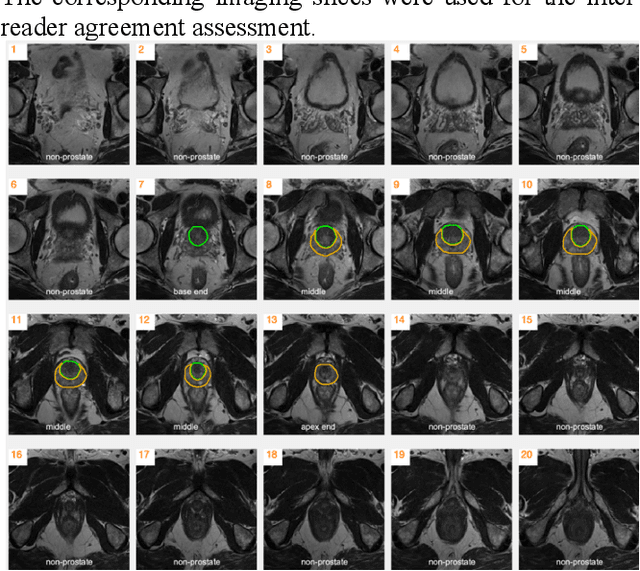
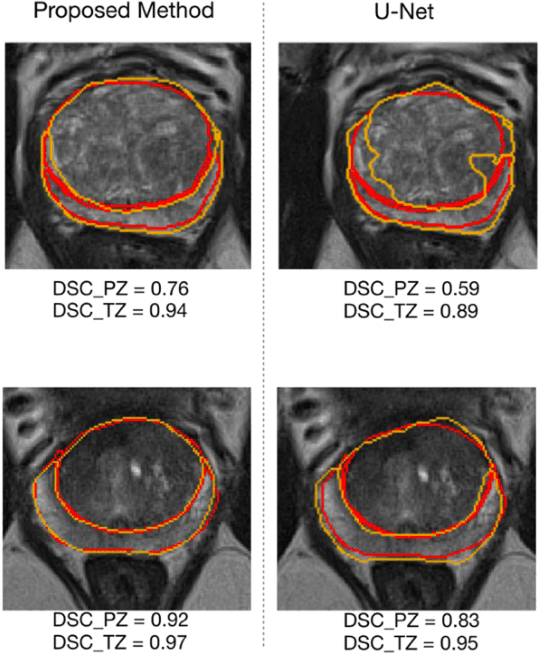
Abstract:Our main objective is to develop a novel deep learning-based algorithm for automatic segmentation of prostate zone and to evaluate the proposed algorithm on an additional independent testing data in comparison with inter-reader consistency between two experts. With IRB approval and HIPAA compliance, we designed a novel convolutional neural network (CNN) for automatic segmentation of the prostatic transition zone (TZ) and peripheral zone (PZ) on T2-weighted (T2w) MRI. The total study cohort included 359 patients from two sources; 313 from a deidentified publicly available dataset (SPIE-AAPM-NCI PROSTATEX challenge) and 46 from a large U.S. tertiary referral center with 3T MRI (external testing dataset (ETD)). The TZ and PZ contours were manually annotated by research fellows, supervised by genitourinary (GU) radiologists. The model was developed using 250 patients and tested internally using the remaining 63 patients from the PROSTATEX (internal testing dataset (ITD)) and tested again (n=46) externally using the ETD. The Dice Similarity Coefficient (DSC) was used to evaluate the segmentation performance. DSCs for PZ and TZ were 0.74 and 0.86 in the ITD respectively. In the ETD, DSCs for PZ and TZ were 0.74 and 0.792, respectively. The inter-reader consistency (Expert 2 vs. Expert 1) were 0.71 (PZ) and 0.75 (TZ). This novel DL algorithm enabled automatic segmentation of PZ and TZ with high accuracy on both ITD and ETD without a performance difference for PZ and less than 10% TZ difference. In the ETD, the proposed method can be comparable to experts in the segmentation of prostate zones.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge