Xiaobo Lai
ME-Net: Multi-Encoder Net Framework for Brain Tumor Segmentation
Mar 21, 2022Abstract:Glioma is the most common and aggressive brain tumor. Magnetic resonance imaging (MRI) plays a vital role to evaluate tumors for the arrangement of tumor surgery and the treatment of subsequent procedures. However, the manual segmentation of the MRI image is strenuous, which limits its clinical application. With the development of deep learning, a large number of automatic segmentation methods have been developed, but most of them stay in 2D images, which leads to subpar performance. Moreover, the serious voxel imbalance between the brain tumor and the background as well as the different sizes and locations of the brain tumor makes the segmentation of 3D images a challenging problem. Aiming at segmenting 3D MRI, we propose a model for brain tumor segmentation with multiple encoders. The structure contains four encoders and one decoder. The four encoders correspond to the four modalities of the MRI image, perform one-to-one feature extraction, and then merge the feature maps of the four modalities into the decoder. This method reduces the difficulty of feature extraction and greatly improves model performance. We also introduced a new loss function named "Categorical Dice", and set different weights for different segmented regions at the same time, which solved the problem of voxel imbalance. We evaluated our approach using the online BraTS 2020 Challenge verification. Our proposed method can achieve promising results in the validation set compared to the state-of-the-art approaches with Dice scores of 0.70249, 0.88267, and 0.73864 for the intact tumor, tumor core, and enhanced tumor, respectively.
3D AGSE-VNet: An Automatic Brain Tumor MRI Data Segmentation Framework
Jul 26, 2021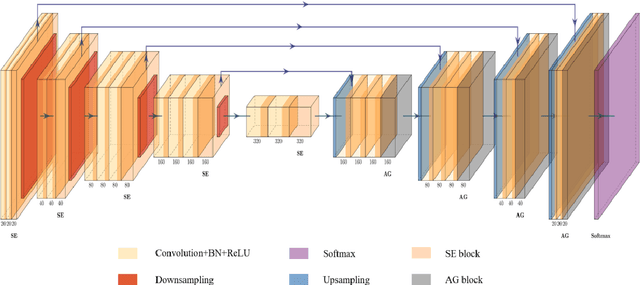
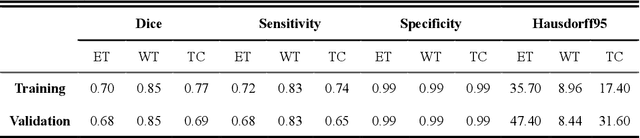
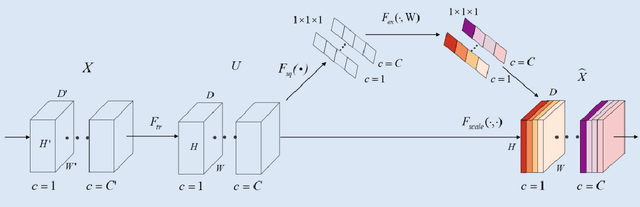
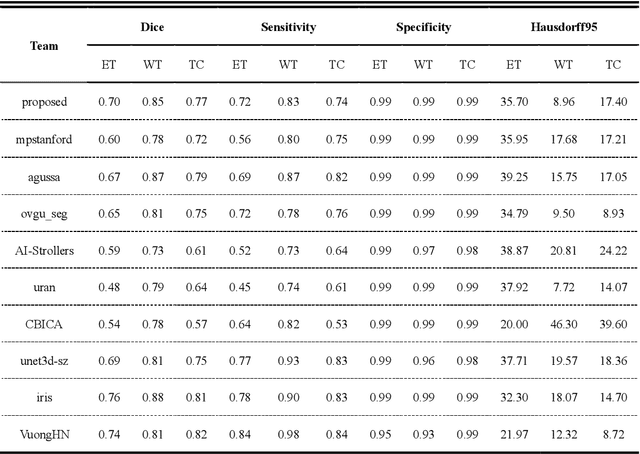
Abstract:Background: Glioma is the most common brain malignant tumor, with a high morbidity rate and a mortality rate of more than three percent, which seriously endangers human health. The main method of acquiring brain tumors in the clinic is MRI. Segmentation of brain tumor regions from multi-modal MRI scan images is helpful for treatment inspection, post-diagnosis monitoring, and effect evaluation of patients. However, the common operation in clinical brain tumor segmentation is still manual segmentation, lead to its time-consuming and large performance difference between different operators, a consistent and accurate automatic segmentation method is urgently needed. Methods: To meet the above challenges, we propose an automatic brain tumor MRI data segmentation framework which is called AGSE-VNet. In our study, the Squeeze and Excite (SE) module is added to each encoder, the Attention Guide Filter (AG) module is added to each decoder, using the channel relationship to automatically enhance the useful information in the channel to suppress the useless information, and use the attention mechanism to guide the edge information and remove the influence of irrelevant information such as noise. Results: We used the BraTS2020 challenge online verification tool to evaluate our approach. The focus of verification is that the Dice scores of the whole tumor (WT), tumor core (TC) and enhanced tumor (ET) are 0.68, 0.85 and 0.70, respectively. Conclusion: Although MRI images have different intensities, AGSE-VNet is not affected by the size of the tumor, and can more accurately extract the features of the three regions, it has achieved impressive results and made outstanding contributions to the clinical diagnosis and treatment of brain tumor patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge