Richard Frayne
Cumming School of Medicine, Department of Radiology, University of Calgary, Calgary, Canada
Trustworthy and Responsible AI for Human-Centric Autonomous Decision-Making Systems
Sep 02, 2024Abstract:Artificial Intelligence (AI) has paved the way for revolutionary decision-making processes, which if harnessed appropriately, can contribute to advancements in various sectors, from healthcare to economics. However, its black box nature presents significant ethical challenges related to bias and transparency. AI applications are hugely impacted by biases, presenting inconsistent and unreliable findings, leading to significant costs and consequences, highlighting and perpetuating inequalities and unequal access to resources. Hence, developing safe, reliable, ethical, and Trustworthy AI systems is essential. Our team of researchers working with Trustworthy and Responsible AI, part of the Transdisciplinary Scholarship Initiative within the University of Calgary, conducts research on Trustworthy and Responsible AI, including fairness, bias mitigation, reproducibility, generalization, interpretability, and authenticity. In this paper, we review and discuss the intricacies of AI biases, definitions, methods of detection and mitigation, and metrics for evaluating bias. We also discuss open challenges with regard to the trustworthiness and widespread application of AI across diverse domains of human-centric decision making, as well as guidelines to foster Responsible and Trustworthy AI models.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
SF2Former: Amyotrophic Lateral Sclerosis Identification From Multi-center MRI Data Using Spatial and Frequency Fusion Transformer
Feb 28, 2023



Abstract:Amyotrophic Lateral Sclerosis (ALS) is a complex neurodegenerative disorder involving motor neuron degeneration. Significant research has begun to establish brain magnetic resonance imaging (MRI) as a potential biomarker to diagnose and monitor the state of the disease. Deep learning has turned into a prominent class of machine learning programs in computer vision and has been successfully employed to solve diverse medical image analysis tasks. However, deep learning-based methods applied to neuroimaging have not achieved superior performance in ALS patients classification from healthy controls due to having insignificant structural changes correlated with pathological features. Therefore, the critical challenge in deep models is to determine useful discriminative features with limited training data. By exploiting the long-range relationship of image features, this study introduces a framework named SF2Former that leverages vision transformer architecture's power to distinguish the ALS subjects from the control group. To further improve the network's performance, spatial and frequency domain information are combined because MRI scans are captured in the frequency domain before being converted to the spatial domain. The proposed framework is trained with a set of consecutive coronal 2D slices, which uses the pre-trained weights on ImageNet by leveraging transfer learning. Finally, a majority voting scheme has been employed to those coronal slices of a particular subject to produce the final classification decision. Our proposed architecture has been thoroughly assessed with multi-modal neuroimaging data using two well-organized versions of the Canadian ALS Neuroimaging Consortium (CALSNIC) multi-center datasets. The experimental results demonstrate the superiority of our proposed strategy in terms of classification accuracy compared with several popular deep learning-based techniques.
Multi-channel MR Reconstruction (MC-MRRec) Challenge -- Comparing Accelerated MR Reconstruction Models and Assessing Their Genereralizability to Datasets Collected with Different Coils
Nov 10, 2020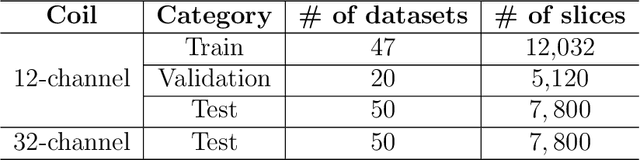
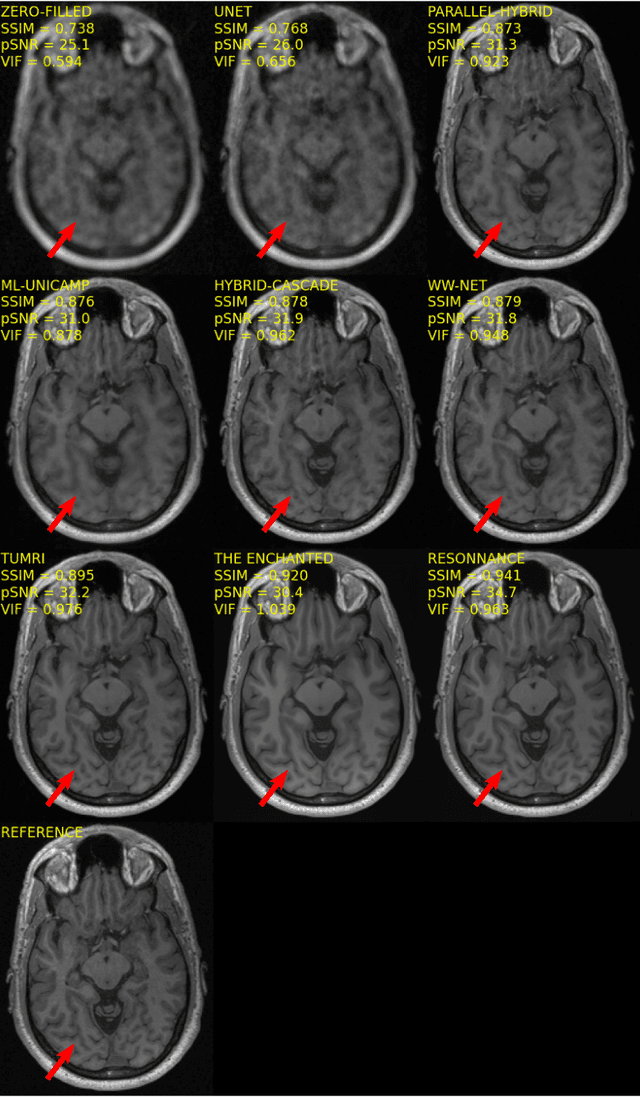
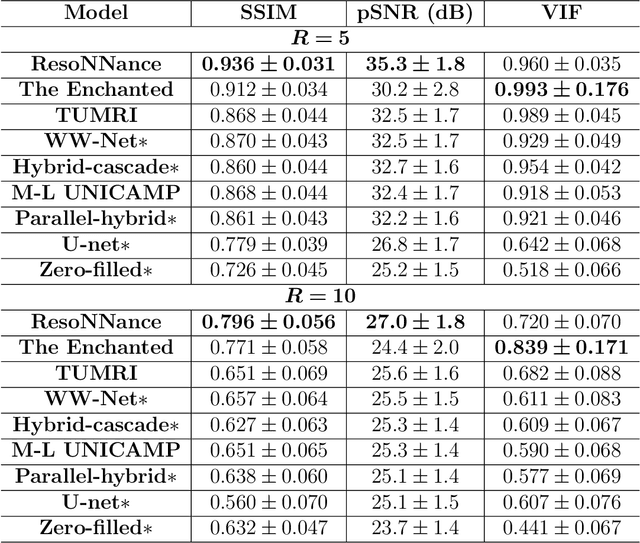
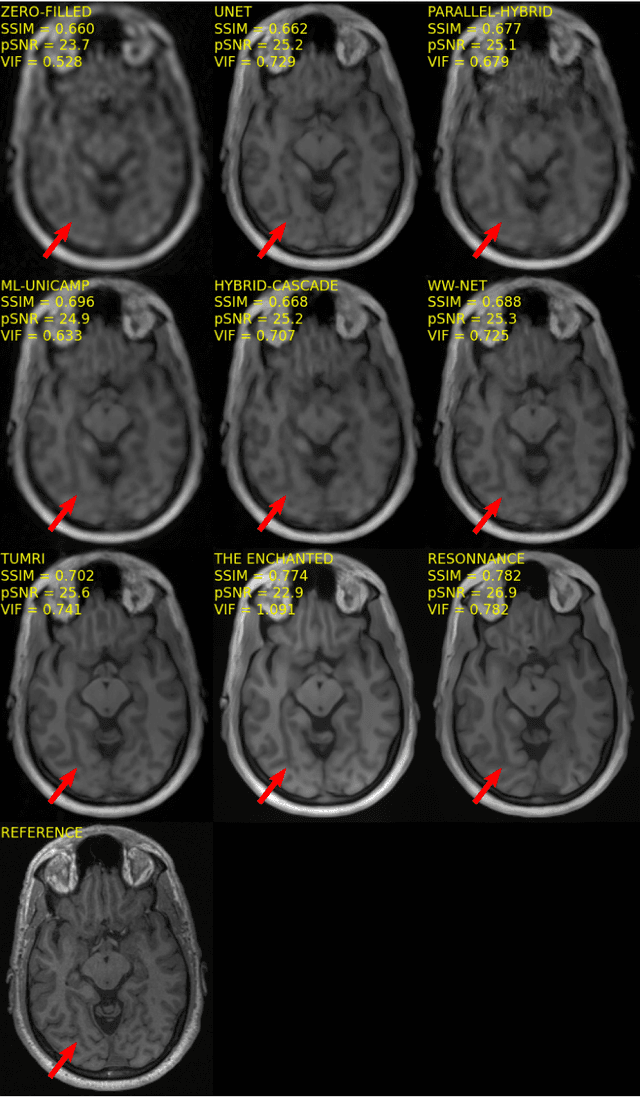
Abstract:The 2020 Multi-channel Magnetic Resonance Reconstruction (MC-MRRec) Challenge had two primary goals: 1) compare different MR image reconstruction models on a large dataset and 2) assess the generalizability of these models to datasets acquired with a different number of receiver coils (i.e., multiple channels). The challenge had two tracks: Track 01 focused on assessing models trained and tested with 12-channel data. Track 02 focused on assessing models trained with 12-channel data and tested on both 12-channel and 32-channel data. While the challenge is ongoing, here we describe the first edition of the challenge and summarise submissions received prior to 5 September 2020. Track 01 had five baseline models and received four independent submissions. Track 02 had two baseline models and received two independent submissions. This manuscript provides relevant comparative information on the current state-of-the-art of MR reconstruction and highlights the challenges of obtaining generalizable models that are required prior to clinical adoption. Both challenge tracks remain open and will provide an objective performance assessment for future submissions. Subsequent editions of the challenge are proposed to investigate new concepts and strategies, such as the integration of potentially available longitudinal information during the MR reconstruction process. An outline of the proposed second edition of the challenge is presented in this manuscript.
Dual-domain Cascade of U-nets for Multi-channel Magnetic Resonance Image Reconstruction
Nov 04, 2019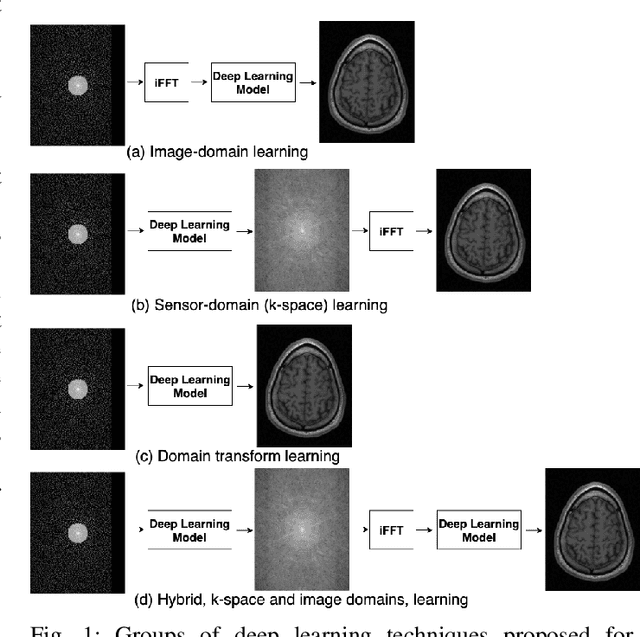
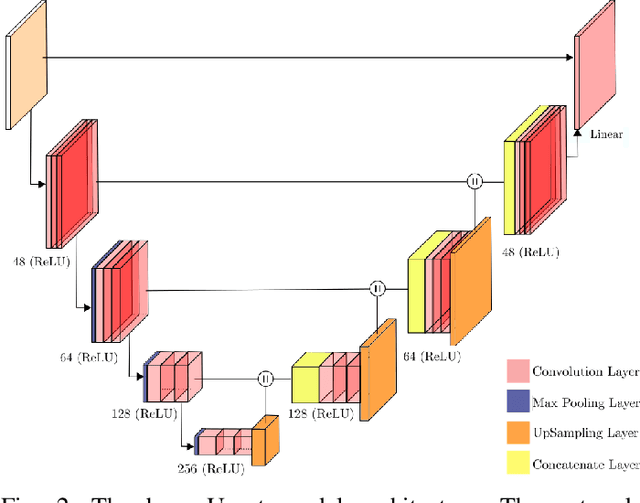
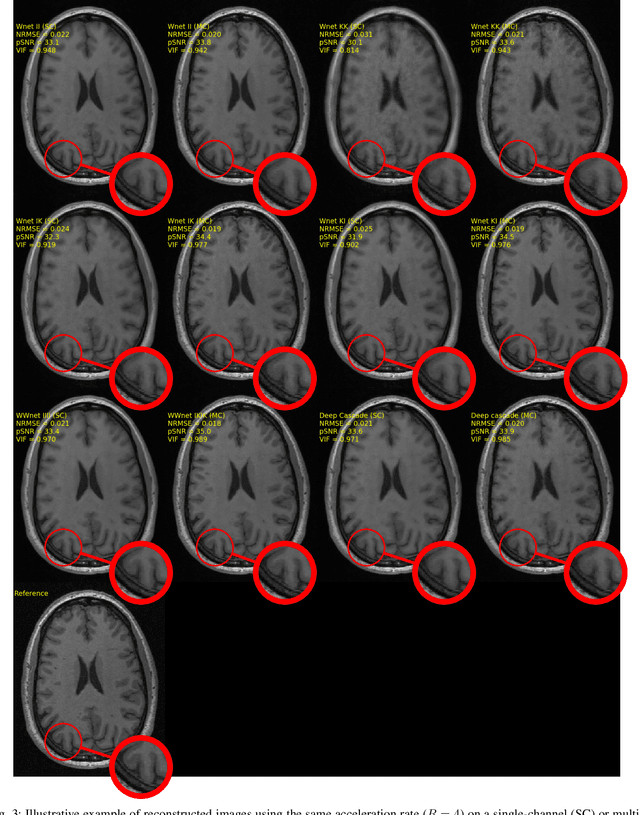
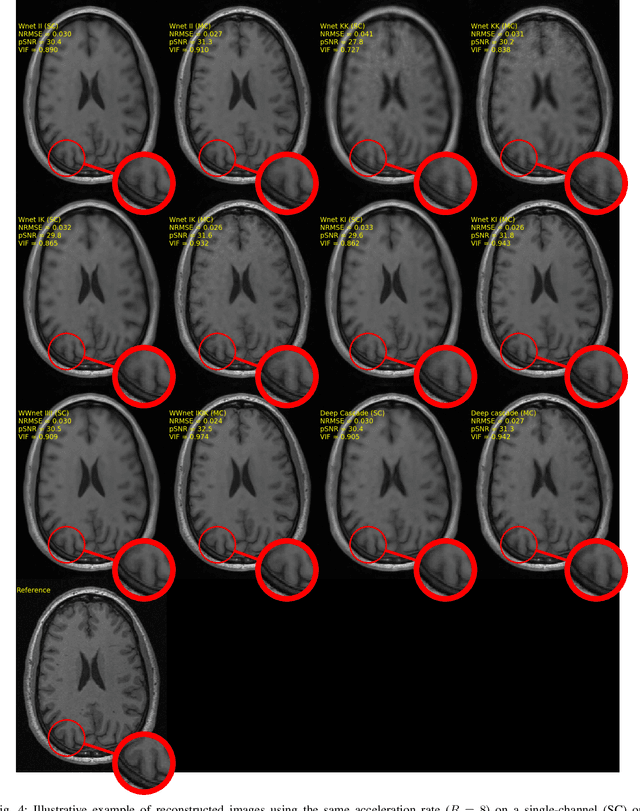
Abstract:The U-net is a deep-learning network model that has been used to solve a number of inverse problems. In this work, the concatenation of two-element U-nets, termed the W-net, operating in k-space (K) and image (I) domains, were evaluated for multi-channel magnetic resonance (MR) image reconstruction. The two element network combinations were evaluated for the four possible image-k-space domain configurations: a) W-net II, b) W-net KK, c) W-net IK, and d) W-net KI were evaluated. Selected promising four element networks (WW-nets) were also examined. Two configurations of each network were compared: 1) Each coil channel processed independently, and 2) all channels processed simultaneously. One hundred and eleven volumetric, T1-weighted, 12-channel coil k-space datasets were used in the experiments. Normalized root mean squared error, peak signal to noise ratio, visual information fidelity and visual inspection were used to assess the reconstructed images against the fully sampled reference images. Our results indicated that networks that operate solely in the image domain are better suited when processing individual channels of multi-channel data independently. Dual domain methods are more advantageous when simultaneously reconstructing all channels of multi-channel data. Also, the appropriate cascade of U-nets compared favorably (p < 0.01) to the previously published, state-of-the-art Deep Cascade model in in three out of four experiments.
A Hybrid Frequency-domain/Image-domain Deep Network for Magnetic Resonance Image Reconstruction
Oct 30, 2018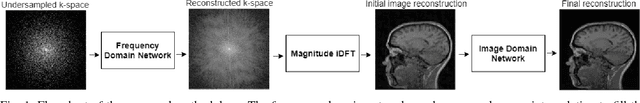
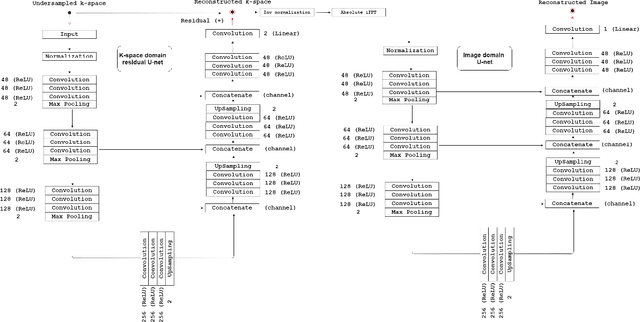
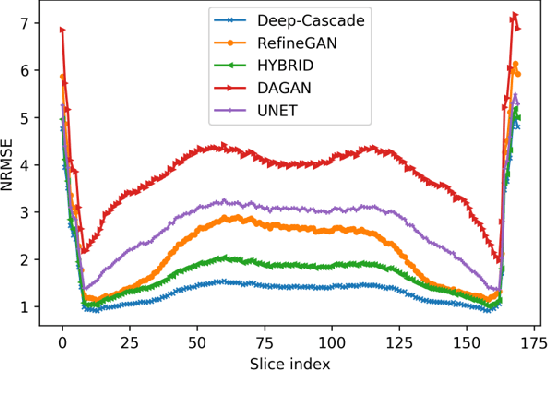
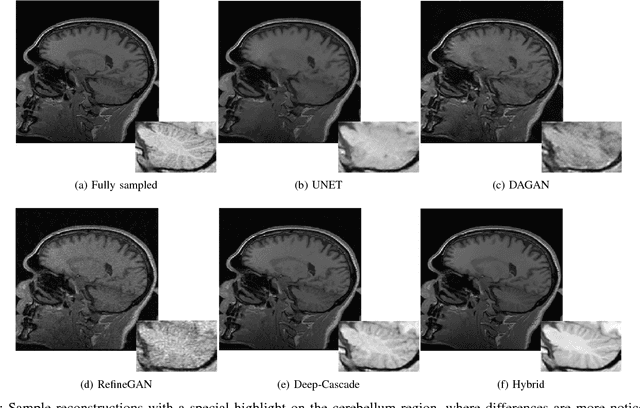
Abstract:Decreasing magnetic resonance (MR) image acquisition times can potentially reduce procedural cost and make MR examinations more accessible. Compressed sensing (CS)-based image reconstruction methods, for example, decrease MR acquisition time by reconstructing high-quality images from data that were originally sampled at rates inferior to the Nyquist-Shannon sampling theorem. In this work we propose a hybrid architecture that works both in the k-space (or frequency-domain) and the image (or spatial) domains. Our network is composed of a complex-valued residual U-net in the k-space domain, an inverse Fast Fourier Transform (iFFT) operation, and a real-valued U-net in the image domain. Our experiments demonstrated, using MR raw k-space data, that the proposed hybrid approach can potentially improve CS reconstruction compared to deep-learning networks that operate only in the image domain. In this study we compare our method with four previously published deep neural networks and examine their ability to reconstruct images that are subsequently used to generate regional volume estimates. We evaluated undersampling ratios of 75% and 80%. Our technique was ranked second in the quantitative analysis, but qualitative analysis indicated that our reconstruction performed the best in hard to reconstruct regions, such as the cerebellum. All images reconstructed with our method were successfully post-processed, and showed good volumetry agreement compared with the fully sampled reconstruction measures.
Convolutional Neural Networks for Skull-stripping in Brain MR Imaging using Consensus-based Silver standard Masks
Apr 13, 2018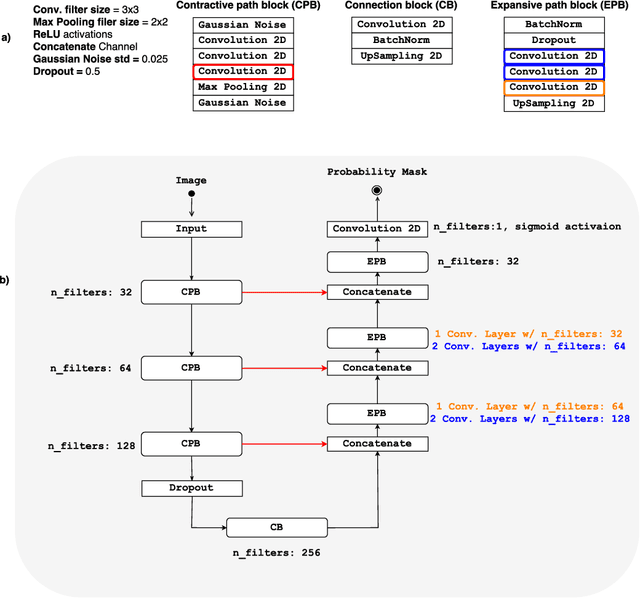
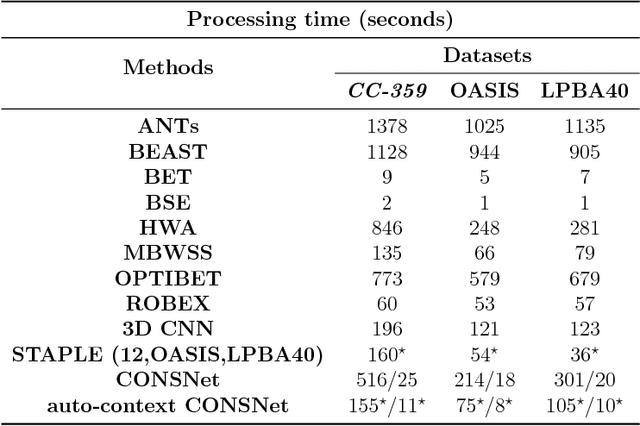
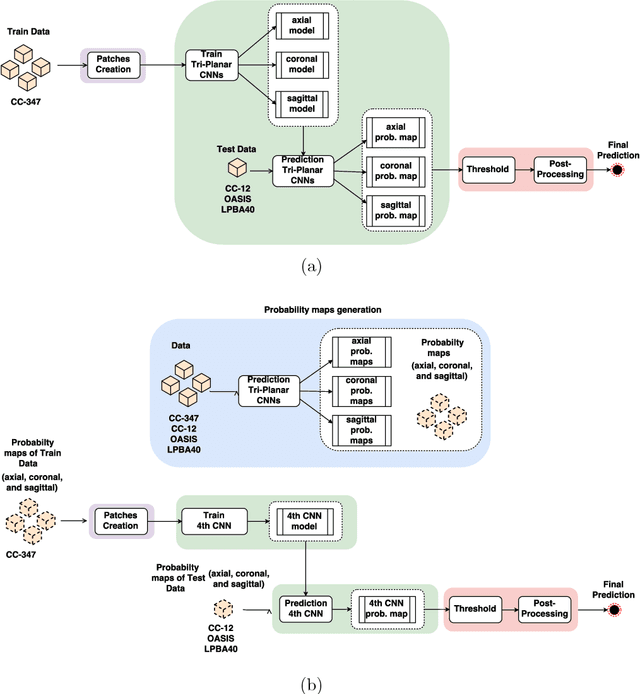
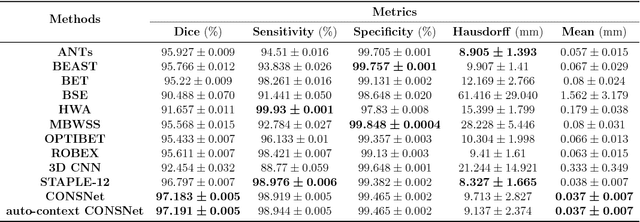
Abstract:Convolutional neural networks (CNN) for medical imaging are constrained by the number of annotated data required in the training stage. Usually, manual annotation is considered to be the "gold standard". However, medical imaging datasets that include expert manual segmentation are scarce as this step is time-consuming, and therefore expensive. Moreover, single-rater manual annotation is most often used in data-driven approaches making the network optimal with respect to only that single expert. In this work, we propose a CNN for brain extraction in magnetic resonance (MR) imaging, that is fully trained with what we refer to as silver standard masks. Our method consists of 1) developing a dataset with "silver standard" masks as input, and implementing both 2) a tri-planar method using parallel 2D U-Net-based CNNs (referred to as CONSNet) and 3) an auto-context implementation of CONSNet. The term CONSNet refers to our integrated approach, i.e., training with silver standard masks and using a 2D U-Net-based architecture. Our results showed that we outperformed (i.e., larger Dice coefficients) the current state-of-the-art SS methods. Our use of silver standard masks reduced the cost of manual annotation, decreased inter-intra-rater variability, and avoided CNN segmentation super-specialization towards one specific manual annotation guideline that can occur when gold standard masks are used. Moreover, the usage of silver standard masks greatly enlarges the volume of input annotated data because we can relatively easily generate labels for unlabeled data. In addition, our method has the advantage that, once trained, it takes only a few seconds to process a typical brain image volume using modern hardware, such as a high-end graphics processing unit. In contrast, many of the other competitive methods have processing times in the order of minutes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge