Convolutional Neural Networks for Skull-stripping in Brain MR Imaging using Consensus-based Silver standard Masks
Paper and Code
Apr 13, 2018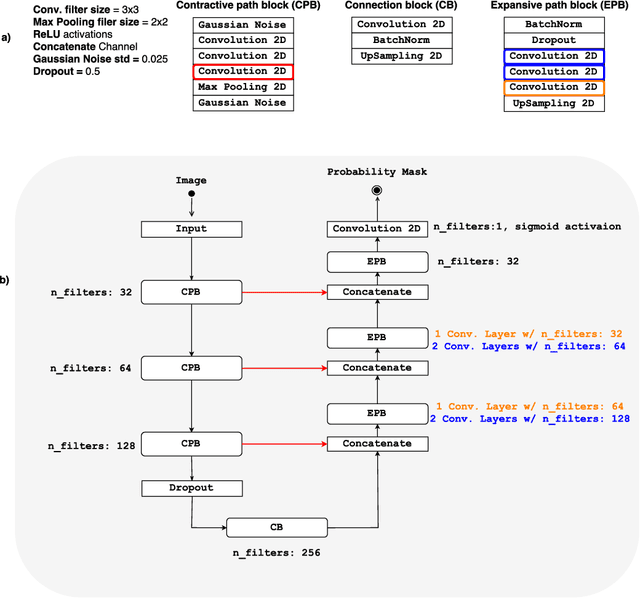
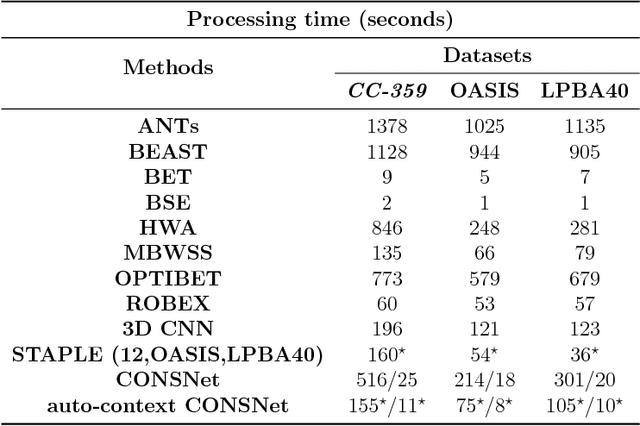
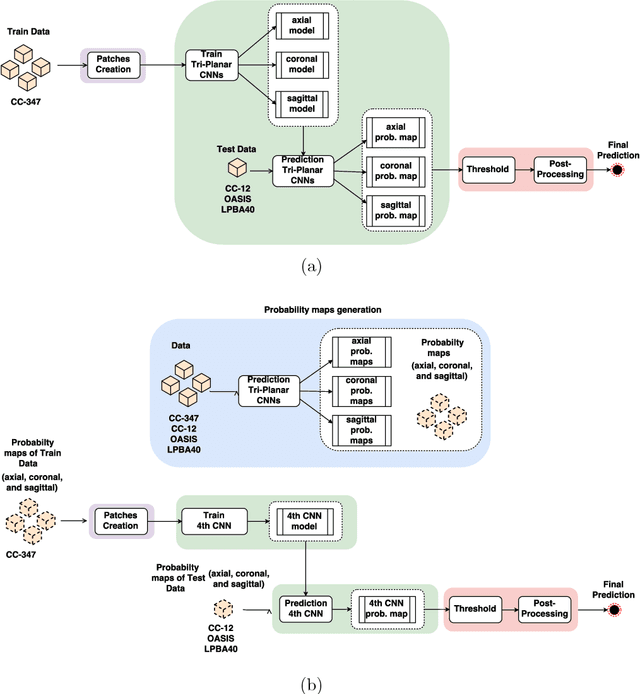
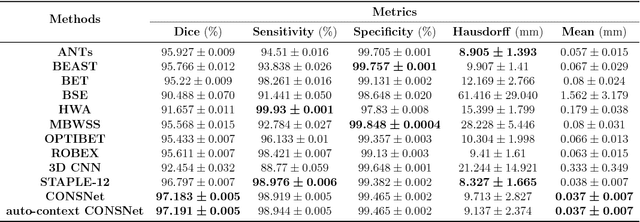
Convolutional neural networks (CNN) for medical imaging are constrained by the number of annotated data required in the training stage. Usually, manual annotation is considered to be the "gold standard". However, medical imaging datasets that include expert manual segmentation are scarce as this step is time-consuming, and therefore expensive. Moreover, single-rater manual annotation is most often used in data-driven approaches making the network optimal with respect to only that single expert. In this work, we propose a CNN for brain extraction in magnetic resonance (MR) imaging, that is fully trained with what we refer to as silver standard masks. Our method consists of 1) developing a dataset with "silver standard" masks as input, and implementing both 2) a tri-planar method using parallel 2D U-Net-based CNNs (referred to as CONSNet) and 3) an auto-context implementation of CONSNet. The term CONSNet refers to our integrated approach, i.e., training with silver standard masks and using a 2D U-Net-based architecture. Our results showed that we outperformed (i.e., larger Dice coefficients) the current state-of-the-art SS methods. Our use of silver standard masks reduced the cost of manual annotation, decreased inter-intra-rater variability, and avoided CNN segmentation super-specialization towards one specific manual annotation guideline that can occur when gold standard masks are used. Moreover, the usage of silver standard masks greatly enlarges the volume of input annotated data because we can relatively easily generate labels for unlabeled data. In addition, our method has the advantage that, once trained, it takes only a few seconds to process a typical brain image volume using modern hardware, such as a high-end graphics processing unit. In contrast, many of the other competitive methods have processing times in the order of minutes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge