Roxana Daneshjou
Stanford University
Visual concept ranking uncovers medical shortcuts used by large multimodal models
Feb 04, 2026Abstract:Ensuring the reliability of machine learning models in safety-critical domains such as healthcare requires auditing methods that can uncover model shortcomings. We introduce a method for identifying important visual concepts within large multimodal models (LMMs) and use it to investigate the behaviors these models exhibit when prompted with medical tasks. We primarily focus on the task of classifying malignant skin lesions from clinical dermatology images, with supplemental experiments including both chest radiographs and natural images. After showing how LMMs display unexpected gaps in performance between different demographic subgroups when prompted with demonstrating examples, we apply our method, Visual Concept Ranking (VCR), to these models and prompts. VCR generates hypotheses related to different visual feature dependencies, which we are then able to validate with manual interventions.
ReasonEdit: Editing Vision-Language Models using Human Reasoning
Feb 03, 2026Abstract:Model editing aims to correct errors in large, pretrained models without altering unrelated behaviors. While some recent works have edited vision-language models (VLMs), no existing editors tackle reasoning-heavy tasks, which typically require humans and models to reason about images. We therefore propose ReasonEdit, the first VLM editor to let users explain their reasoning during editing, introducing a new, practical model editing setup. ReasonEdit continuously stores human reasoning in a codebook, and retrieves only relevant facts during inference using a novel topology-balanced multimodal embedding method inspired by network science. Across four VLMs on multiple rationale-based visual question answering datasets, ReasonEdit achieves state-of-the-art editing performance, ultimately showing that using human reasoning during editing greatly improves edit generalization.
Explainable AI as a Double-Edged Sword in Dermatology: The Impact on Clinicians versus The Public
Dec 14, 2025Abstract:Artificial intelligence (AI) is increasingly permeating healthcare, from physician assistants to consumer applications. Since AI algorithm's opacity challenges human interaction, explainable AI (XAI) addresses this by providing AI decision-making insight, but evidence suggests XAI can paradoxically induce over-reliance or bias. We present results from two large-scale experiments (623 lay people; 153 primary care physicians, PCPs) combining a fairness-based diagnosis AI model and different XAI explanations to examine how XAI assistance, particularly multimodal large language models (LLMs), influences diagnostic performance. AI assistance balanced across skin tones improved accuracy and reduced diagnostic disparities. However, LLM explanations yielded divergent effects: lay users showed higher automation bias - accuracy boosted when AI was correct, reduced when AI erred - while experienced PCPs remained resilient, benefiting irrespective of AI accuracy. Presenting AI suggestions first also led to worse outcomes when the AI was incorrect for both groups. These findings highlight XAI's varying impact based on expertise and timing, underscoring LLMs as a "double-edged sword" in medical AI and informing future human-AI collaborative system design.
Prompt Triage: Structured Optimization Enhances Vision-Language Model Performance on Medical Imaging Benchmarks
Nov 14, 2025Abstract:Vision-language foundation models (VLMs) show promise for diverse imaging tasks but often underperform on medical benchmarks. Prior efforts to improve performance include model finetuning, which requires large domain-specific datasets and significant compute, or manual prompt engineering, which is hard to generalize and often inaccessible to medical institutions seeking to deploy these tools. These challenges motivate interest in approaches that draw on a model's embedded knowledge while abstracting away dependence on human-designed prompts to enable scalable, weight-agnostic performance improvements. To explore this, we adapt the Declarative Self-improving Python (DSPy) framework for structured automated prompt optimization in medical vision-language systems through a comprehensive, formal evaluation. We implement prompting pipelines for five medical imaging tasks across radiology, gastroenterology, and dermatology, evaluating 10 open-source VLMs with four prompt optimization techniques. Optimized pipelines achieved a median relative improvement of 53% over zero-shot prompting baselines, with the largest gains ranging from 300% to 3,400% on tasks where zero-shot performance is low. These results highlight the substantial potential of applying automated prompt optimization to medical AI systems, demonstrating significant gains for vision-based applications requiring accurate clinical image interpretation. By reducing dependence on prompt design to elicit intended outputs, these techniques allow clinicians to focus on patient care and clinical decision-making. Furthermore, our experiments offer scalability and preserve data privacy, demonstrating performance improvement on open-source VLMs. We publicly release our evaluation pipelines to support reproducible research on specialized medical tasks, available at https://github.com/DaneshjouLab/prompt-triage-lab.
MedHELM: Holistic Evaluation of Large Language Models for Medical Tasks
May 26, 2025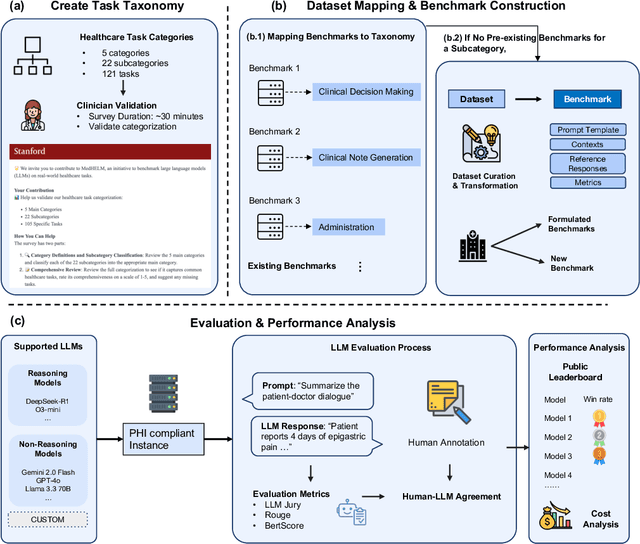
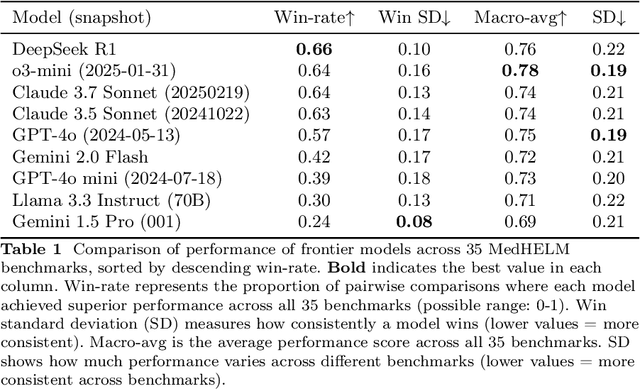
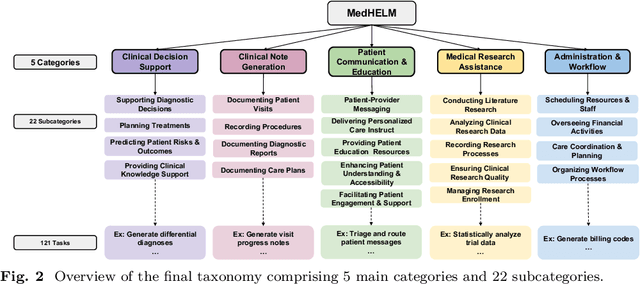
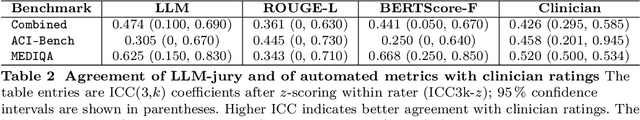
Abstract:While large language models (LLMs) achieve near-perfect scores on medical licensing exams, these evaluations inadequately reflect the complexity and diversity of real-world clinical practice. We introduce MedHELM, an extensible evaluation framework for assessing LLM performance for medical tasks with three key contributions. First, a clinician-validated taxonomy spanning 5 categories, 22 subcategories, and 121 tasks developed with 29 clinicians. Second, a comprehensive benchmark suite comprising 35 benchmarks (17 existing, 18 newly formulated) providing complete coverage of all categories and subcategories in the taxonomy. Third, a systematic comparison of LLMs with improved evaluation methods (using an LLM-jury) and a cost-performance analysis. Evaluation of 9 frontier LLMs, using the 35 benchmarks, revealed significant performance variation. Advanced reasoning models (DeepSeek R1: 66% win-rate; o3-mini: 64% win-rate) demonstrated superior performance, though Claude 3.5 Sonnet achieved comparable results at 40% lower estimated computational cost. On a normalized accuracy scale (0-1), most models performed strongly in Clinical Note Generation (0.73-0.85) and Patient Communication & Education (0.78-0.83), moderately in Medical Research Assistance (0.65-0.75), and generally lower in Clinical Decision Support (0.56-0.72) and Administration & Workflow (0.53-0.63). Our LLM-jury evaluation method achieved good agreement with clinician ratings (ICC = 0.47), surpassing both average clinician-clinician agreement (ICC = 0.43) and automated baselines including ROUGE-L (0.36) and BERTScore-F1 (0.44). Claude 3.5 Sonnet achieved comparable performance to top models at lower estimated cost. These findings highlight the importance of real-world, task-specific evaluation for medical use of LLMs and provides an open source framework to enable this.
BiasICL: In-Context Learning and Demographic Biases of Vision Language Models
Mar 04, 2025



Abstract:Vision language models (VLMs) show promise in medical diagnosis, but their performance across demographic subgroups when using in-context learning (ICL) remains poorly understood. We examine how the demographic composition of demonstration examples affects VLM performance in two medical imaging tasks: skin lesion malignancy prediction and pneumothorax detection from chest radiographs. Our analysis reveals that ICL influences model predictions through multiple mechanisms: (1) ICL allows VLMs to learn subgroup-specific disease base rates from prompts and (2) ICL leads VLMs to make predictions that perform differently across demographic groups, even after controlling for subgroup-specific disease base rates. Our empirical results inform best-practices for prompting current VLMs (specifically examining demographic subgroup performance, and matching base rates of labels to target distribution at a bulk level and within subgroups), while also suggesting next steps for improving our theoretical understanding of these models.
SycEval: Evaluating LLM Sycophancy
Feb 12, 2025



Abstract:Large language models (LLMs) are increasingly applied in educational, clinical, and professional settings, but their tendency for sycophancy -- prioritizing user agreement over independent reasoning -- poses risks to reliability. This study introduces a framework to evaluate sycophantic behavior in ChatGPT-4o, Claude-Sonnet, and Gemini-1.5-Pro across AMPS (mathematics) and MedQuad (medical advice) datasets. Sycophantic behavior was observed in 58.19% of cases, with Gemini exhibiting the highest rate (62.47%) and ChatGPT the lowest (56.71%). Progressive sycophancy, leading to correct answers, occurred in 43.52% of cases, while regressive sycophancy, leading to incorrect answers, was observed in 14.66%. Preemptive rebuttals demonstrated significantly higher sycophancy rates than in-context rebuttals (61.75% vs. 56.52%, $Z=5.87$, $p<0.001$), particularly in computational tasks, where regressive sycophancy increased significantly (preemptive: 8.13%, in-context: 3.54%, $p<0.001$). Simple rebuttals maximized progressive sycophancy ($Z=6.59$, $p<0.001$), while citation-based rebuttals exhibited the highest regressive rates ($Z=6.59$, $p<0.001$). Sycophantic behavior showed high persistence (78.5%, 95% CI: [77.2%, 79.8%]) regardless of context or model. These findings emphasize the risks and opportunities of deploying LLMs in structured and dynamic domains, offering insights into prompt programming and model optimization for safer AI applications.
Best Practices for Large Language Models in Radiology
Dec 02, 2024



Abstract:At the heart of radiological practice is the challenge of integrating complex imaging data with clinical information to produce actionable insights. Nuanced application of language is key for various activities, including managing requests, describing and interpreting imaging findings in the context of clinical data, and concisely documenting and communicating the outcomes. The emergence of large language models (LLMs) offers an opportunity to improve the management and interpretation of the vast data in radiology. Despite being primarily general-purpose, these advanced computational models demonstrate impressive capabilities in specialized language-related tasks, even without specific training. Unlocking the potential of LLMs for radiology requires basic understanding of their foundations and a strategic approach to navigate their idiosyncrasies. This review, drawing from practical radiology and machine learning expertise and recent literature, provides readers insight into the potential of LLMs in radiology. It examines best practices that have so far stood the test of time in the rapidly evolving landscape of LLMs. This includes practical advice for optimizing LLM characteristics for radiology practices along with limitations, effective prompting, and fine-tuning strategies.
Almanac Copilot: Towards Autonomous Electronic Health Record Navigation
May 14, 2024



Abstract:Clinicians spend large amounts of time on clinical documentation, and inefficiencies impact quality of care and increase clinician burnout. Despite the promise of electronic medical records (EMR), the transition from paper-based records has been negatively associated with clinician wellness, in part due to poor user experience, increased burden of documentation, and alert fatigue. In this study, we present Almanac Copilot, an autonomous agent capable of assisting clinicians with EMR-specific tasks such as information retrieval and order placement. On EHR-QA, a synthetic evaluation dataset of 300 common EHR queries based on real patient data, Almanac Copilot obtains a successful task completion rate of 74% (n = 221 tasks) with a mean score of 2.45 over 3 (95% CI:2.34-2.56). By automating routine tasks and streamlining the documentation process, our findings highlight the significant potential of autonomous agents to mitigate the cognitive load imposed on clinicians by current EMR systems.
Assessing The Potential Of Mid-Sized Language Models For Clinical QA
Apr 24, 2024



Abstract:Large language models, such as GPT-4 and Med-PaLM, have shown impressive performance on clinical tasks; however, they require access to compute, are closed-source, and cannot be deployed on device. Mid-size models such as BioGPT-large, BioMedLM, LLaMA 2, and Mistral 7B avoid these drawbacks, but their capacity for clinical tasks has been understudied. To help assess their potential for clinical use and help researchers decide which model they should use, we compare their performance on two clinical question-answering (QA) tasks: MedQA and consumer query answering. We find that Mistral 7B is the best performing model, winning on all benchmarks and outperforming models trained specifically for the biomedical domain. While Mistral 7B's MedQA score of 63.0% approaches the original Med-PaLM, and it often can produce plausible responses to consumer health queries, room for improvement still exists. This study provides the first head-to-head assessment of open source mid-sized models on clinical tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge