Oliver Aalami
Medicine on the Edge: Comparative Performance Analysis of On-Device LLMs for Clinical Reasoning
Feb 13, 2025Abstract:The deployment of Large Language Models (LLM) on mobile devices offers significant potential for medical applications, enhancing privacy, security, and cost-efficiency by eliminating reliance on cloud-based services and keeping sensitive health data local. However, the performance and accuracy of on-device LLMs in real-world medical contexts remain underexplored. In this study, we benchmark publicly available on-device LLMs using the AMEGA dataset, evaluating accuracy, computational efficiency, and thermal limitation across various mobile devices. Our results indicate that compact general-purpose models like Phi-3 Mini achieve a strong balance between speed and accuracy, while medically fine-tuned models such as Med42 and Aloe attain the highest accuracy. Notably, deploying LLMs on older devices remains feasible, with memory constraints posing a greater challenge than raw processing power. Our study underscores the potential of on-device LLMs for healthcare while emphasizing the need for more efficient inference and models tailored to real-world clinical reasoning.
Dynamic Fog Computing for Enhanced LLM Execution in Medical Applications
Aug 08, 2024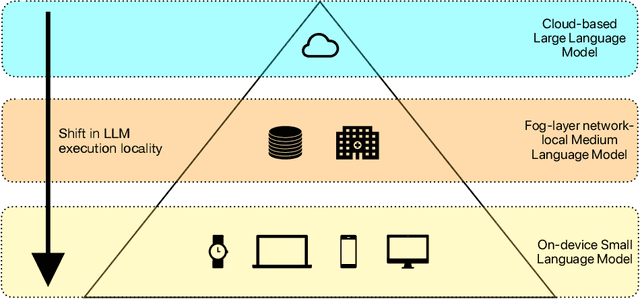

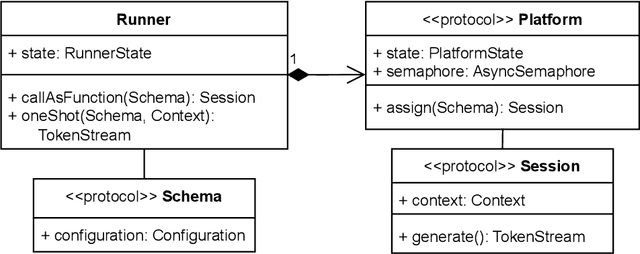
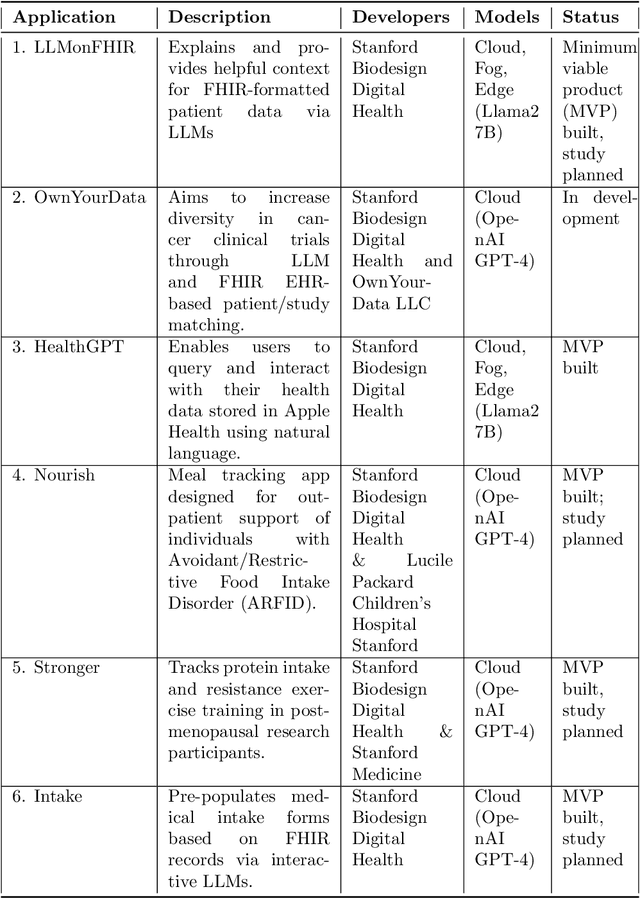
Abstract:The ability of large language models (LLMs) to transform, interpret, and comprehend vast quantities of heterogeneous data presents a significant opportunity to enhance data-driven care delivery. However, the sensitive nature of protected health information (PHI) raises valid concerns about data privacy and trust in remote LLM platforms. In addition, the cost associated with cloud-based artificial intelligence (AI) services continues to impede widespread adoption. To address these challenges, we propose a shift in the LLM execution environment from opaque, centralized cloud providers to a decentralized and dynamic fog computing architecture. By executing open-weight LLMs in more trusted environments, such as the user's edge device or a fog layer within a local network, we aim to mitigate the privacy, trust, and financial challenges associated with cloud-based LLMs. We further present SpeziLLM, an open-source framework designed to facilitate rapid and seamless leveraging of different LLM execution layers and lowering barriers to LLM integration in digital health applications. We demonstrate SpeziLLM's broad applicability across six digital health applications, showcasing its versatility in various healthcare settings.
Almanac Copilot: Towards Autonomous Electronic Health Record Navigation
May 14, 2024



Abstract:Clinicians spend large amounts of time on clinical documentation, and inefficiencies impact quality of care and increase clinician burnout. Despite the promise of electronic medical records (EMR), the transition from paper-based records has been negatively associated with clinician wellness, in part due to poor user experience, increased burden of documentation, and alert fatigue. In this study, we present Almanac Copilot, an autonomous agent capable of assisting clinicians with EMR-specific tasks such as information retrieval and order placement. On EHR-QA, a synthetic evaluation dataset of 300 common EHR queries based on real patient data, Almanac Copilot obtains a successful task completion rate of 74% (n = 221 tasks) with a mean score of 2.45 over 3 (95% CI:2.34-2.56). By automating routine tasks and streamlining the documentation process, our findings highlight the significant potential of autonomous agents to mitigate the cognitive load imposed on clinicians by current EMR systems.
IMIL: Interactive Medical Image Learning Framework
Apr 17, 2024



Abstract:Data augmentations are widely used in training medical image deep learning models to increase the diversity and size of sparse datasets. However, commonly used augmentation techniques can result in loss of clinically relevant information from medical images, leading to incorrect predictions at inference time. We propose the Interactive Medical Image Learning (IMIL) framework, a novel approach for improving the training of medical image analysis algorithms that enables clinician-guided intermediate training data augmentations on misprediction outliers, focusing the algorithm on relevant visual information. To prevent the model from using irrelevant features during training, IMIL will 'blackout' clinician-designated irrelevant regions and replace the original images with the augmented samples. This ensures that for originally mispredicted samples, the algorithm subsequently attends only to relevant regions and correctly correlates them with the respective diagnosis. We validate the efficacy of IMIL using radiology residents and compare its performance to state-of-the-art data augmentations. A 4.2% improvement in accuracy over ResNet-50 was observed when using IMIL on only 4% of the training set. Our study demonstrates the utility of clinician-guided interactive training to achieve meaningful data augmentations for medical image analysis algorithms.
LLM on FHIR -- Demystifying Health Records
Jan 25, 2024Abstract:Objective: To enhance health literacy and accessibility of health information for a diverse patient population by developing a patient-centered artificial intelligence (AI) solution using large language models (LLMs) and Fast Healthcare Interoperability Resources (FHIR) application programming interfaces (APIs). Materials and Methods: The research involved developing LLM on FHIR, an open-source mobile application allowing users to interact with their health records using LLMs. The app is built on Stanford's Spezi ecosystem and uses OpenAI's GPT-4. A pilot study was conducted with the SyntheticMass patient dataset and evaluated by medical experts to assess the app's effectiveness in increasing health literacy. The evaluation focused on the accuracy, relevance, and understandability of the LLM's responses to common patient questions. Results: LLM on FHIR demonstrated varying but generally high degrees of accuracy and relevance in providing understandable health information to patients. The app effectively translated medical data into patient-friendly language and was able to adapt its responses to different patient profiles. However, challenges included variability in LLM responses and the need for precise filtering of health data. Discussion and Conclusion: LLMs offer significant potential in improving health literacy and making health records more accessible. LLM on FHIR, as a pioneering application in this field, demonstrates the feasibility and challenges of integrating LLMs into patient care. While promising, the implementation and pilot also highlight risks such as inconsistent responses and the importance of replicable output. Future directions include better resource identification mechanisms and executing LLMs on-device to enhance privacy and reduce costs.
Studying the Impact of Augmentations on Medical Confidence Calibration
Aug 23, 2023



Abstract:The clinical explainability of convolutional neural networks (CNN) heavily relies on the joint interpretation of a model's predicted diagnostic label and associated confidence. A highly certain or uncertain model can significantly impact clinical decision-making. Thus, ensuring that confidence estimates reflect the true correctness likelihood for a prediction is essential. CNNs are often poorly calibrated and prone to overconfidence leading to improper measures of uncertainty. This creates the need for confidence calibration. However, accuracy and performance-based evaluations of CNNs are commonly used as the sole benchmark for medical tasks. Taking into consideration the risks associated with miscalibration is of high importance. In recent years, modern augmentation techniques, which cut, mix, and combine images, have been introduced. Such augmentations have benefited CNNs through regularization, robustness to adversarial samples, and calibration. Standard augmentations based on image scaling, rotating, and zooming, are widely leveraged in the medical domain to combat the scarcity of data. In this paper, we evaluate the effects of three modern augmentation techniques, CutMix, MixUp, and CutOut on the calibration and performance of CNNs for medical tasks. CutMix improved calibration the most while CutOut often lowered the level of calibration.
Studying the Effects of Self-Attention for Medical Image Analysis
Sep 02, 2021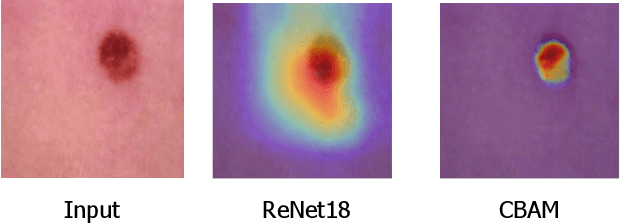
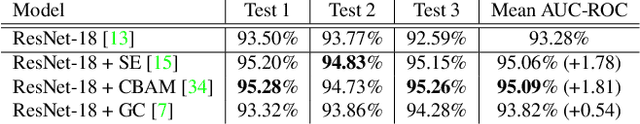
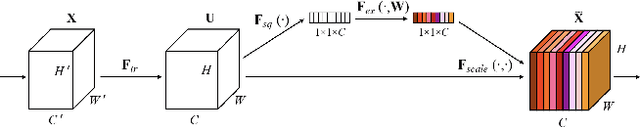
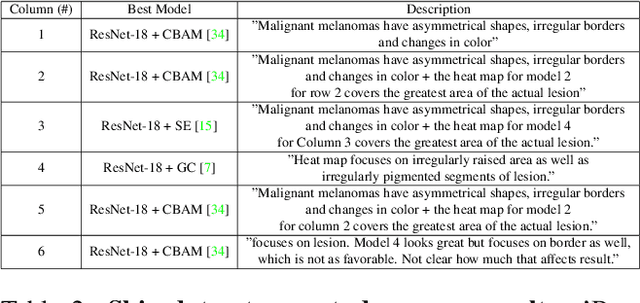
Abstract:When the trained physician interprets medical images, they understand the clinical importance of visual features. By applying cognitive attention, they apply greater focus onto clinically relevant regions while disregarding unnecessary features. The use of computer vision to automate the classification of medical images is widely studied. However, the standard convolutional neural network (CNN) does not necessarily employ subconscious feature relevancy evaluation techniques similar to the trained medical specialist and evaluates features more generally. Self-attention mechanisms enable CNNs to focus more on semantically important regions or aggregated relevant context with long-range dependencies. By using attention, medical image analysis systems can potentially become more robust by focusing on more important clinical feature regions. In this paper, we provide a comprehensive comparison of various state-of-the-art self-attention mechanisms across multiple medical image analysis tasks. Through both quantitative and qualitative evaluations along with a clinical user-centric survey study, we aim to provide a deeper understanding of the effects of self-attention in medical computer vision tasks.
Waveform Phasicity Prediction from Arterial Sounds through Spectogram Analysis using Convolutional Neural Networks for Limb Perfusion Assessment
Apr 22, 2021

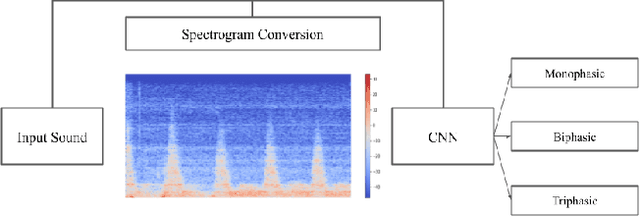
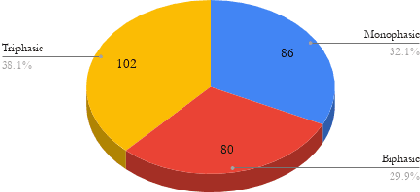
Abstract:Peripheral Arterial Disease (PAD) is a common form of arterial occlusive disease that is challenging to evaluate at the point-of-care. Hand-held dopplers are the most ubiquitous device used to evaluate circulation and allows providers to audibly "listen" to the blood flow. Providers use the audible feedback to subjectively assess whether the sound characteristics are consistent with Monophasic, Biphasic, or Triphasic waveforms. Subjective assessment of doppler sounds raises suspicion of PAD and leads to further testing, often delaying definitive treatment. Misdiagnoses are also possible with subjective interpretation of doppler waveforms. This paper presents a Deep Learning system that has the ability to predict waveform phasicity through analysis of hand-held doppler sounds. We collected 268 four-second recordings on an iPhone taken during a formal vascular lab study in patients with cardiovascular disease. Our end-to-end system works by converting input sound into a spectrogram which visually represents frequency changes in temporal patterns. This conversion enables visual differentiation between the phasicity classes. With these changes present, a custom trained Convolutional Neural Network (CNN) is used for prediction through learned feature extraction. The performance of the model was evaluated via calculation of the F1 score and accuracy metrics. The system received an F1 score of 90.57% and an accuracy of 96.23%. Our Deep Learning system is not computationally expensive and has the ability for integration within several applications. When used in a clinic, this system has the capability of preventing misdiagnosis and gives practitioners a second opinion that can be useful in the evaluation of PAD.
Deepwound: Automated Postoperative Wound Assessment and Surgical Site Surveillance through Convolutional Neural Networks
Jul 11, 2018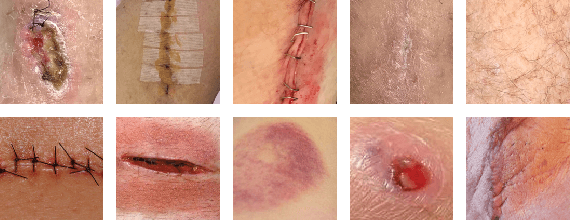
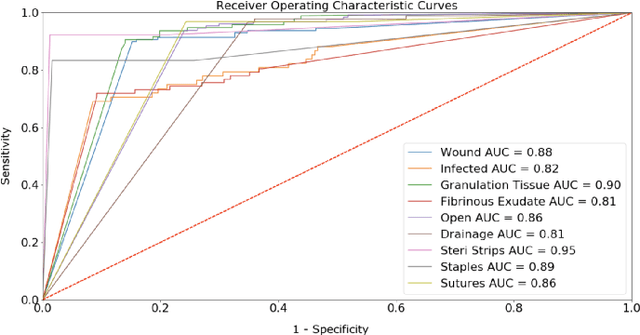


Abstract:Postoperative wound complications are a significant cause of expense for hospitals, doctors, and patients. Hence, an effective method to diagnose the onset of wound complications is strongly desired. Algorithmically classifying wound images is a difficult task due to the variability in the appearance of wound sites. Convolutional neural networks (CNNs), a subgroup of artificial neural networks that have shown great promise in analyzing visual imagery, can be leveraged to categorize surgical wounds. We present a multi-label CNN ensemble, Deepwound, trained to classify wound images using only image pixels and corresponding labels as inputs. Our final computational model can accurately identify the presence of nine labels: drainage, fibrinous exudate, granulation tissue, surgical site infection, open wound, staples, steri strips, and sutures. Our model achieves receiver operating curve (ROC) area under curve (AUC) scores, sensitivity, specificity, and F1 scores superior to prior work in this area. Smartphones provide a means to deliver accessible wound care due to their increasing ubiquity. Paired with deep neural networks, they offer the capability to provide clinical insight to assist surgeons during postoperative care. We also present a mobile application frontend to Deepwound that assists patients in tracking their wound and surgical recovery from the comfort of their home.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge