Allan Halpern
A Patient-Centric Dataset of Images and Metadata for Identifying Melanomas Using Clinical Context
Aug 07, 2020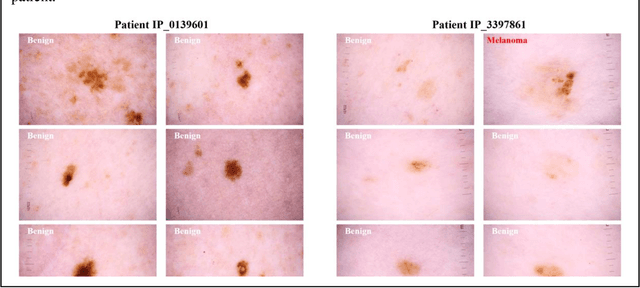
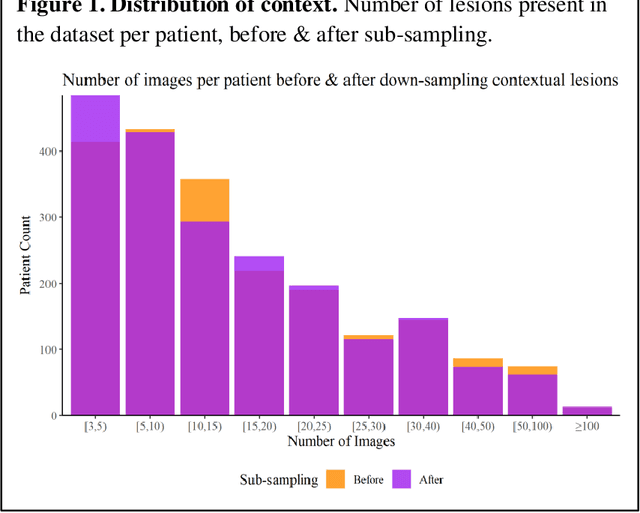
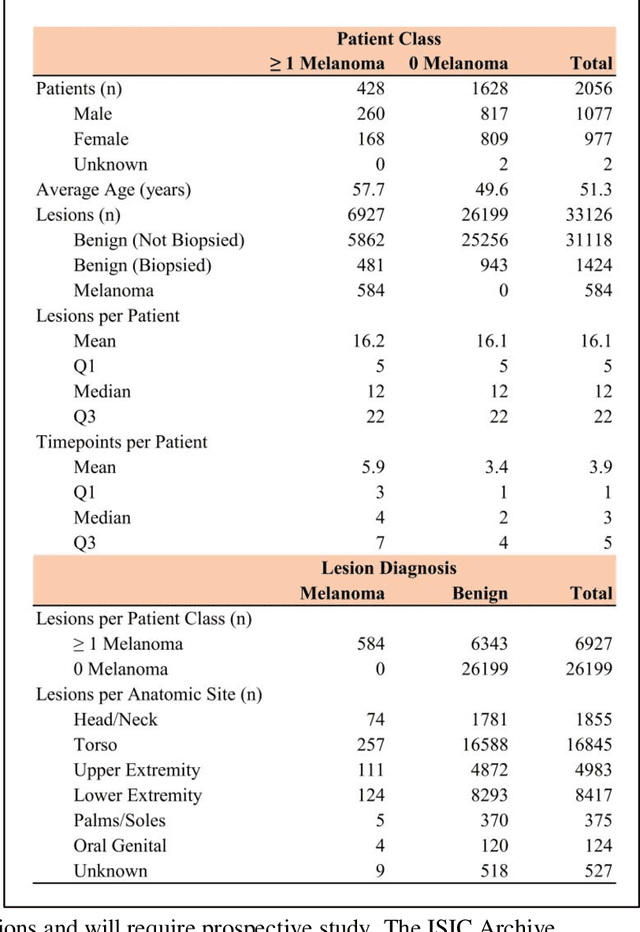
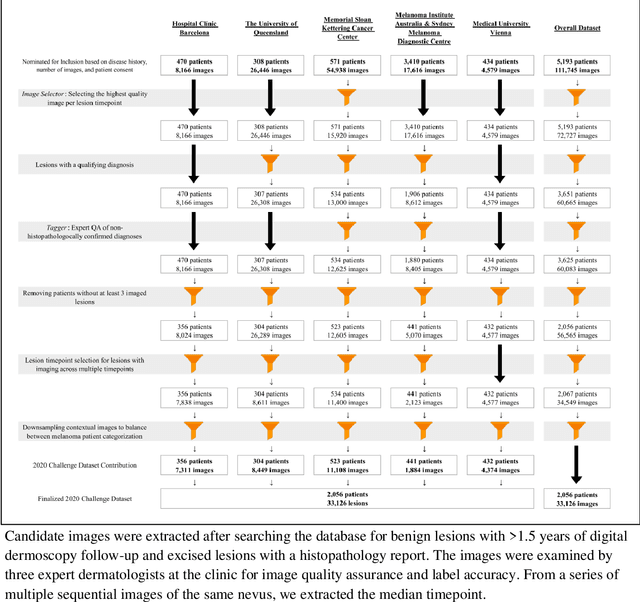
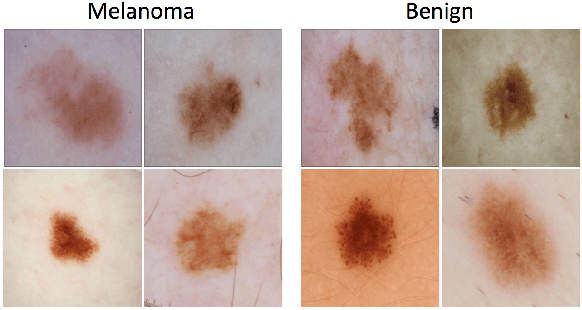
Abstract:Prior skin image datasets have not addressed patient-level information obtained from multiple skin lesions from the same patient. Though artificial intelligence classification algorithms have achieved expert-level performance in controlled studies examining single images, in practice dermatologists base their judgment holistically from multiple lesions on the same patient. The 2020 SIIM-ISIC Melanoma Classification challenge dataset described herein was constructed to address this discrepancy between prior challenges and clinical practice, providing for each image in the dataset an identifier allowing lesions from the same patient to be mapped to one another. This patient-level contextual information is frequently used by clinicians to diagnose melanoma and is especially useful in ruling out false positives in patients with many atypical nevi. The dataset represents 2,056 patients from three continents with an average of 16 lesions per patient, consisting of 33,126 dermoscopic images and 584 histopathologically confirmed melanomas compared with benign melanoma mimickers.
Skin Lesion Analysis Toward Melanoma Detection 2018: A Challenge Hosted by the International Skin Imaging Collaboration (ISIC)
Mar 29, 2019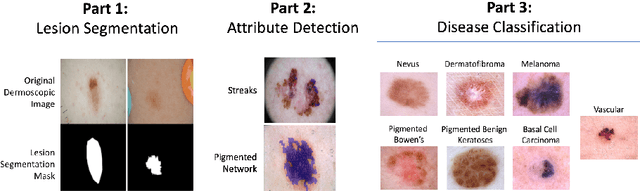
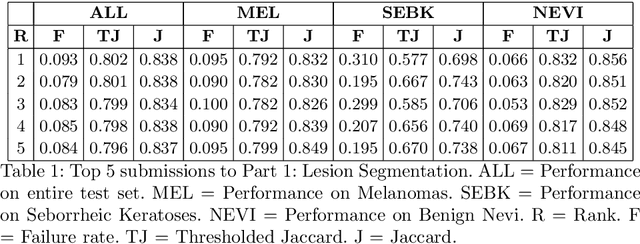
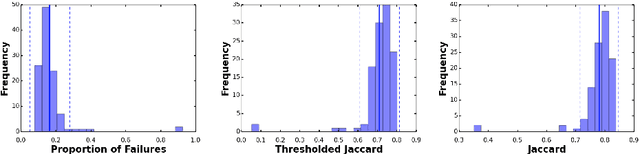
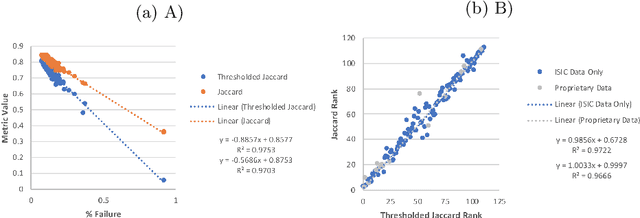
Abstract:This work summarizes the results of the largest skin image analysis challenge in the world, hosted by the International Skin Imaging Collaboration (ISIC), a global partnership that has organized the world's largest public repository of dermoscopic images of skin. The challenge was hosted in 2018 at the Medical Image Computing and Computer Assisted Intervention (MICCAI) conference in Granada, Spain. The dataset included over 12,500 images across 3 tasks. 900 users registered for data download, 115 submitted to the lesion segmentation task, 25 submitted to the lesion attribute detection task, and 159 submitted to the disease classification task. Novel evaluation protocols were established, including a new test for segmentation algorithm performance, and a test for algorithm ability to generalize. Results show that top segmentation algorithms still fail on over 10% of images on average, and algorithms with equal performance on test data can have different abilities to generalize. This is an important consideration for agencies regulating the growing set of machine learning tools in the healthcare domain, and sets a new standard for future public challenges in healthcare.
Collaborative Human-AI (CHAI): Evidence-Based Interpretable Melanoma Classification in Dermoscopic Images
Aug 01, 2018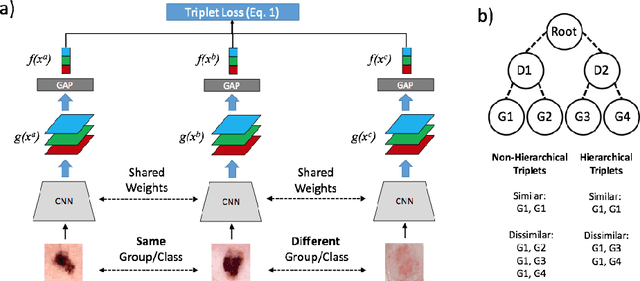
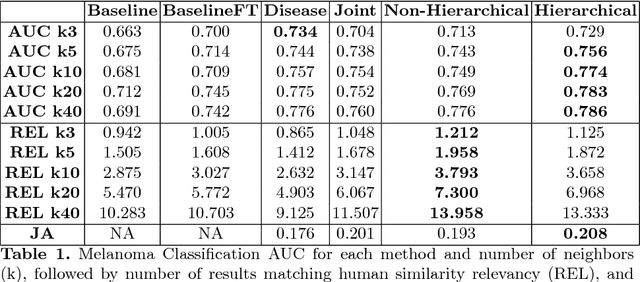
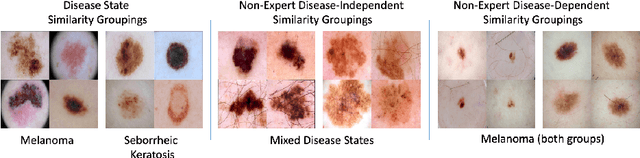
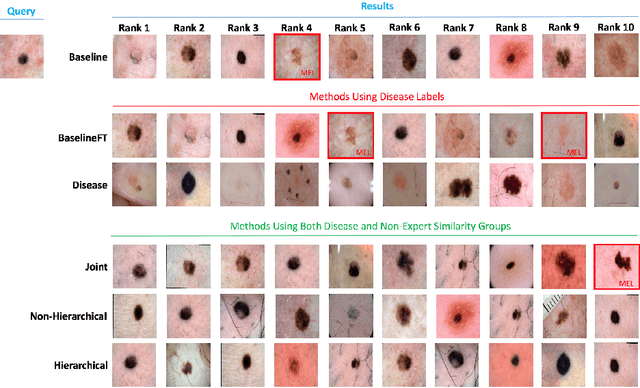
Abstract:Automated dermoscopic image analysis has witnessed rapid growth in diagnostic performance. Yet adoption faces resistance, in part, because no evidence is provided to support decisions. In this work, an approach for evidence-based classification is presented. A feature embedding is learned with CNNs, triplet-loss, and global average pooling, and used to classify via kNN search. Evidence is provided as both the discovered neighbors, as well as localized image regions most relevant to measuring distance between query and neighbors. To ensure that results are relevant in terms of both label accuracy and human visual similarity for any skill level, a novel hierarchical triplet logic is implemented to jointly learn an embedding according to disease labels and non-expert similarity. Results are improved over baselines trained on disease labels alone, as well as standard multiclass loss. Quantitative relevance of results, according to non-expert similarity, as well as localized image regions, are also significantly improved.
Skin Lesion Analysis Toward Melanoma Detection: A Challenge at the 2017 International Symposium on Biomedical Imaging , Hosted by the International Skin Imaging Collaboration
Jan 08, 2018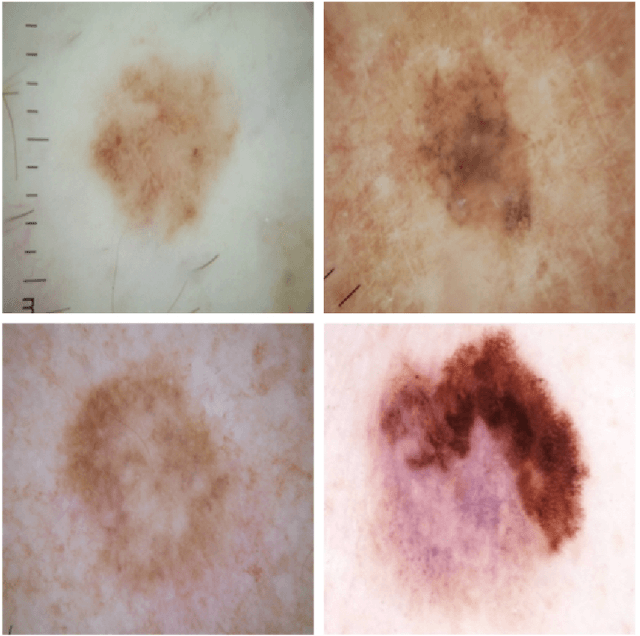
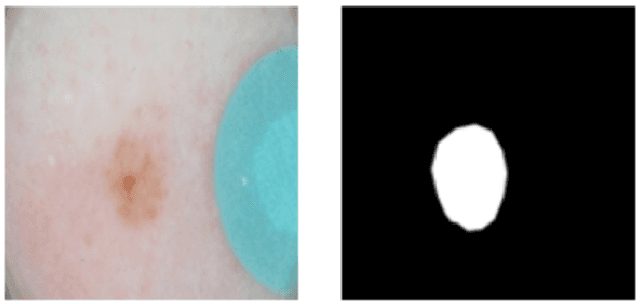
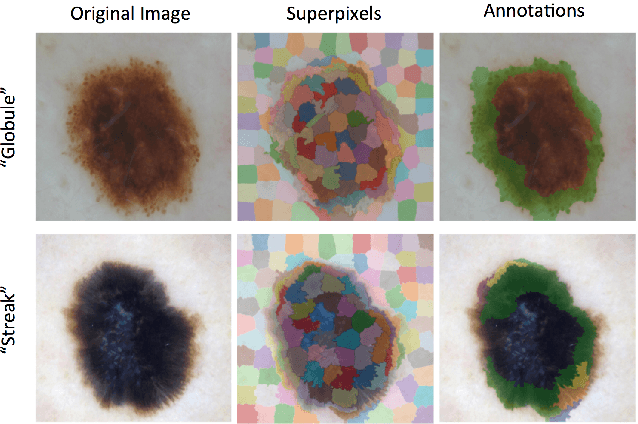
Abstract:This article describes the design, implementation, and results of the latest installment of the dermoscopic image analysis benchmark challenge. The goal is to support research and development of algorithms for automated diagnosis of melanoma, the most lethal skin cancer. The challenge was divided into 3 tasks: lesion segmentation, feature detection, and disease classification. Participation involved 593 registrations, 81 pre-submissions, 46 finalized submissions (including a 4-page manuscript), and approximately 50 attendees, making this the largest standardized and comparative study in this field to date. While the official challenge duration and ranking of participants has concluded, the dataset snapshots remain available for further research and development.
Deep Learning Ensembles for Melanoma Recognition in Dermoscopy Images
Oct 18, 2016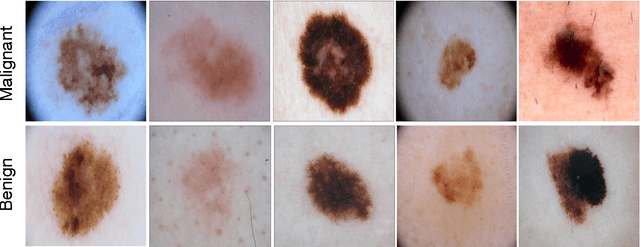

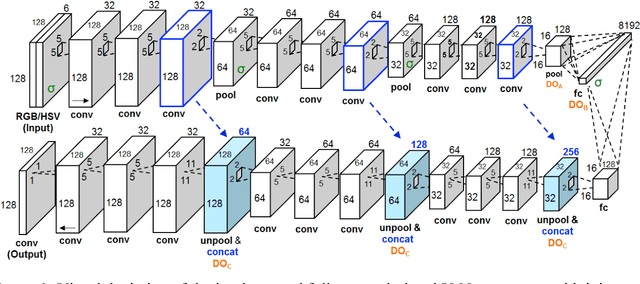
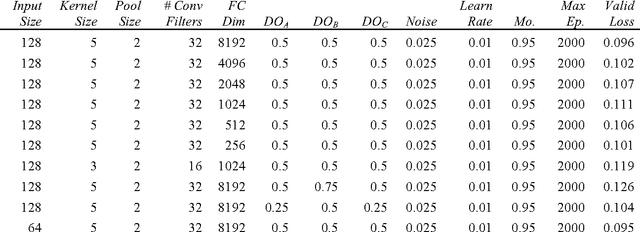
Abstract:Melanoma is the deadliest form of skin cancer. While curable with early detection, only highly trained specialists are capable of accurately recognizing the disease. As expertise is in limited supply, automated systems capable of identifying disease could save lives, reduce unnecessary biopsies, and reduce costs. Toward this goal, we propose a system that combines recent developments in deep learning with established machine learning approaches, creating ensembles of methods that are capable of segmenting skin lesions, as well as analyzing the detected area and surrounding tissue for melanoma detection. The system is evaluated using the largest publicly available benchmark dataset of dermoscopic images, containing 900 training and 379 testing images. New state-of-the-art performance levels are demonstrated, leading to an improvement in the area under receiver operating characteristic curve of 7.5% (0.843 vs. 0.783), in average precision of 4% (0.649 vs. 0.624), and in specificity measured at the clinically relevant 95% sensitivity operating point 2.9 times higher than the previous state-of-the-art (36.8% specificity compared to 12.5%). Compared to the average of 8 expert dermatologists on a subset of 100 test images, the proposed system produces a higher accuracy (76% vs. 70.5%), and specificity (62% vs. 59%) evaluated at an equivalent sensitivity (82%).
* URL for the IBM Journal of Research and Development: http://www.research.ibm.com/journal/
Skin Lesion Analysis toward Melanoma Detection: A Challenge at the International Symposium on Biomedical Imaging 2016, hosted by the International Skin Imaging Collaboration
May 04, 2016



Abstract:In this article, we describe the design and implementation of a publicly accessible dermatology image analysis benchmark challenge. The goal of the challenge is to sup- port research and development of algorithms for automated diagnosis of melanoma, a lethal form of skin cancer, from dermoscopic images. The challenge was divided into sub-challenges for each task involved in image analysis, including lesion segmentation, dermoscopic feature detection within a lesion, and classification of melanoma. Training data included 900 images. A separate test dataset of 379 images was provided to measure resultant performance of systems developed with the training data. Ground truth for both training and test sets was generated by a panel of dermoscopic experts. In total, there were 79 submissions from a group of 38 participants, making this the largest standardized and comparative study for melanoma diagnosis in dermoscopic images to date. While the official challenge duration and ranking of participants has concluded, the datasets remain available for further research and development.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge