Joshua D. Warner
Opportunistic Promptable Segmentation: Leveraging Routine Radiological Annotations to Guide 3D CT Lesion Segmentation
Jan 30, 2026Abstract:The development of machine learning models for CT imaging depends on the availability of large, high-quality, and diverse annotated datasets. Although large volumes of CT images and reports are readily available in clinical picture archiving and communication systems (PACS), 3D segmentations of critical findings are costly to obtain, typically requiring extensive manual annotation by radiologists. On the other hand, it is common for radiologists to provide limited annotations of findings during routine reads, such as line measurements and arrows, that are often stored in PACS as GSPS objects. We posit that these sparse annotations can be extracted along with CT volumes and converted into 3D segmentations using promptable segmentation models, a paradigm we term Opportunistic Promptable Segmentation. To enable this paradigm, we propose SAM2CT, the first promptable segmentation model designed to convert radiologist annotations into 3D segmentations in CT volumes. SAM2CT builds upon SAM2 by extending the prompt encoder to support arrow and line inputs and by introducing Memory-Conditioned Memories (MCM), a memory encoding strategy tailored to 3D medical volumes. On public lesion segmentation benchmarks, SAM2CT outperforms existing promptable segmentation models and similarly trained baselines, achieving Dice similarity coefficients of 0.649 for arrow prompts and 0.757 for line prompts. Applying the model to pre-existing GSPS annotations from a clinical PACS (N = 60), SAM2CT generates 3D segmentations that are clinically acceptable or require only minor adjustments in 87% of cases, as scored by radiologists. Additionally, SAM2CT demonstrates strong zero-shot performance on select Emergency Department findings. These results suggest that large-scale mining of historical GSPS annotations represents a promising and scalable approach for generating 3D CT segmentation datasets.
PETAR: Localized Findings Generation with Mask-Aware Vision-Language Modeling for PET Automated Reporting
Oct 31, 2025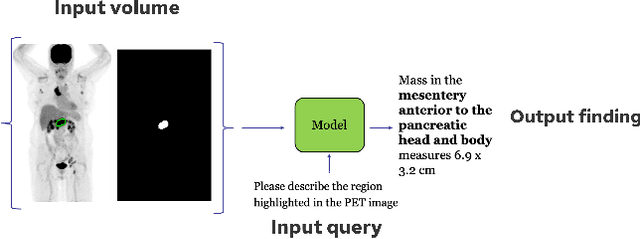
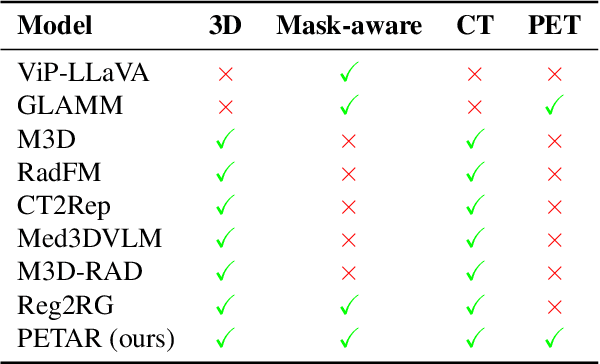
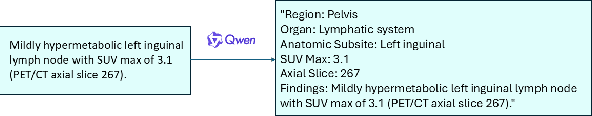
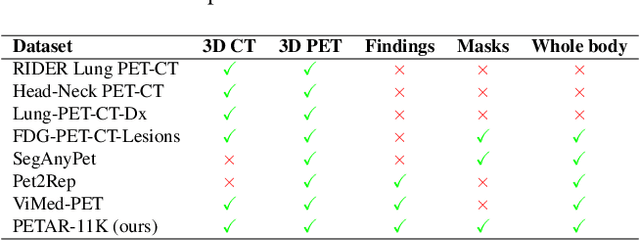
Abstract:Recent advances in vision-language models (VLMs) have enabled impressive multimodal reasoning, yet most medical applications remain limited to 2D imaging. In this work, we extend VLMs to 3D positron emission tomography and computed tomography (PET/CT), a domain characterized by large volumetric data, small and dispersed lesions, and lengthy radiology reports. We introduce a large-scale dataset comprising over 11,000 lesion-level descriptions paired with 3D segmentations from more than 5,000 PET/CT exams, extracted via a hybrid rule-based and large language model (LLM) pipeline. Building upon this dataset, we propose PETAR-4B, a 3D mask-aware vision-language model that integrates PET, CT, and lesion contours for spatially grounded report generation. PETAR bridges global contextual reasoning with fine-grained lesion awareness, producing clinically coherent and localized findings. Comprehensive automated and human evaluations demonstrate that PETAR substantially improves PET/CT report generation quality, advancing 3D medical vision-language understanding.
From Embeddings to Accuracy: Comparing Foundation Models for Radiographic Classification
May 16, 2025Abstract:Foundation models, pretrained on extensive datasets, have significantly advanced machine learning by providing robust and transferable embeddings applicable to various domains, including medical imaging diagnostics. This study evaluates the utility of embeddings derived from both general-purpose and medical domain-specific foundation models for training lightweight adapter models in multi-class radiography classification, focusing specifically on tube placement assessment. A dataset comprising 8842 radiographs classified into seven distinct categories was employed to extract embeddings using six foundation models: DenseNet121, BiomedCLIP, Med-Flamingo, MedImageInsight, Rad-DINO, and CXR-Foundation. Adapter models were subsequently trained using classical machine learning algorithms. Among these combinations, MedImageInsight embeddings paired with an support vector machine adapter yielded the highest mean area under the curve (mAUC) at 93.8%, followed closely by Rad-DINO (91.1%) and CXR-Foundation (89.0%). In comparison, BiomedCLIP and DenseNet121 exhibited moderate performance with mAUC scores of 83.0% and 81.8%, respectively, whereas Med-Flamingo delivered the lowest performance at 75.1%. Notably, most adapter models demonstrated computational efficiency, achieving training within one minute and inference within seconds on CPU, underscoring their practicality for clinical applications. Furthermore, fairness analyses on adapters trained on MedImageInsight-derived embeddings indicated minimal disparities, with gender differences in performance within 2% and standard deviations across age groups not exceeding 3%. These findings confirm that foundation model embeddings-especially those from MedImageInsight-facilitate accurate, computationally efficient, and equitable diagnostic classification using lightweight adapters for radiographic image analysis.
scikit-image: Image processing in Python
Jul 23, 2014



Abstract:scikit-image is an image processing library that implements algorithms and utilities for use in research, education and industry applications. It is released under the liberal "Modified BSD" open source license, provides a well-documented API in the Python programming language, and is developed by an active, international team of collaborators. In this paper we highlight the advantages of open source to achieve the goals of the scikit-image library, and we showcase several real-world image processing applications that use scikit-image.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge