Changhee Lee
PETAR: Localized Findings Generation with Mask-Aware Vision-Language Modeling for PET Automated Reporting
Oct 31, 2025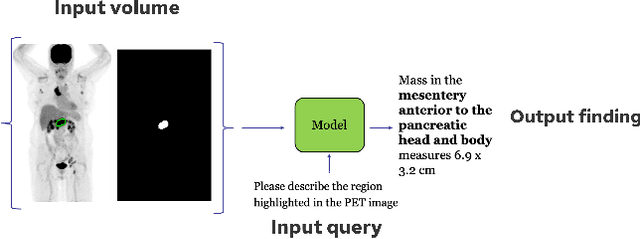
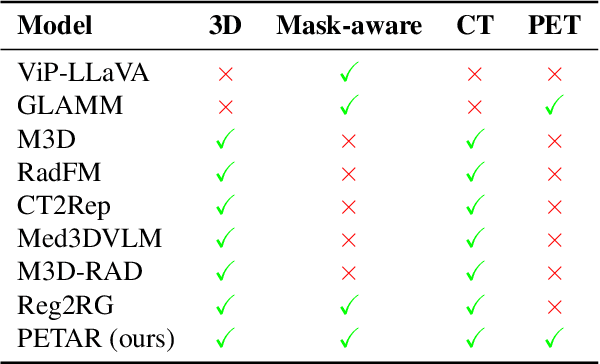
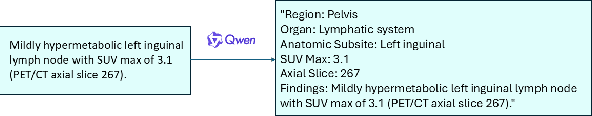
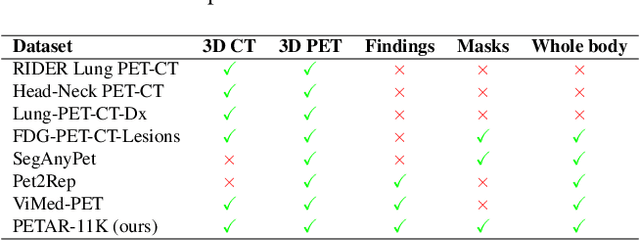
Abstract:Recent advances in vision-language models (VLMs) have enabled impressive multimodal reasoning, yet most medical applications remain limited to 2D imaging. In this work, we extend VLMs to 3D positron emission tomography and computed tomography (PET/CT), a domain characterized by large volumetric data, small and dispersed lesions, and lengthy radiology reports. We introduce a large-scale dataset comprising over 11,000 lesion-level descriptions paired with 3D segmentations from more than 5,000 PET/CT exams, extracted via a hybrid rule-based and large language model (LLM) pipeline. Building upon this dataset, we propose PETAR-4B, a 3D mask-aware vision-language model that integrates PET, CT, and lesion contours for spatially grounded report generation. PETAR bridges global contextual reasoning with fine-grained lesion awareness, producing clinically coherent and localized findings. Comprehensive automated and human evaluations demonstrate that PETAR substantially improves PET/CT report generation quality, advancing 3D medical vision-language understanding.
BioBridge: Unified Bio-Embedding with Bridging Modality in Code-Switched EMR
Dec 16, 2024
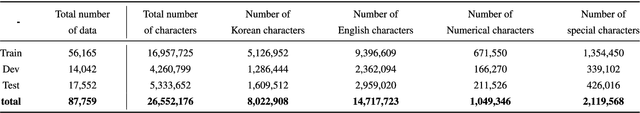
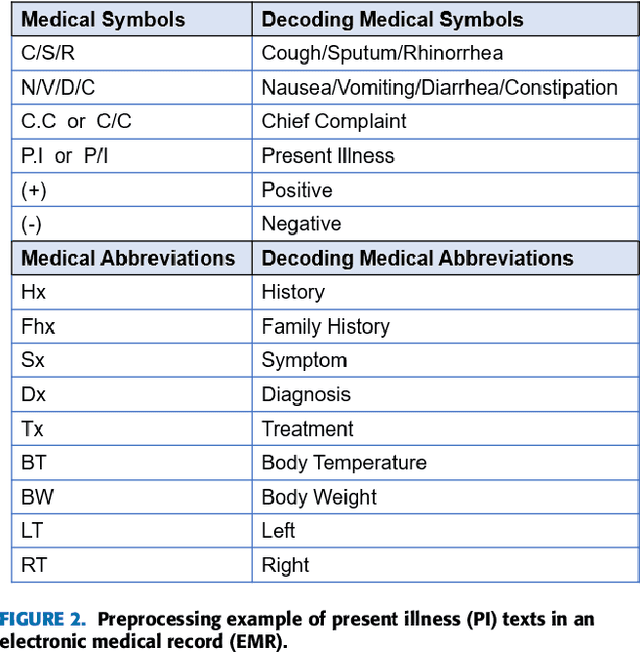
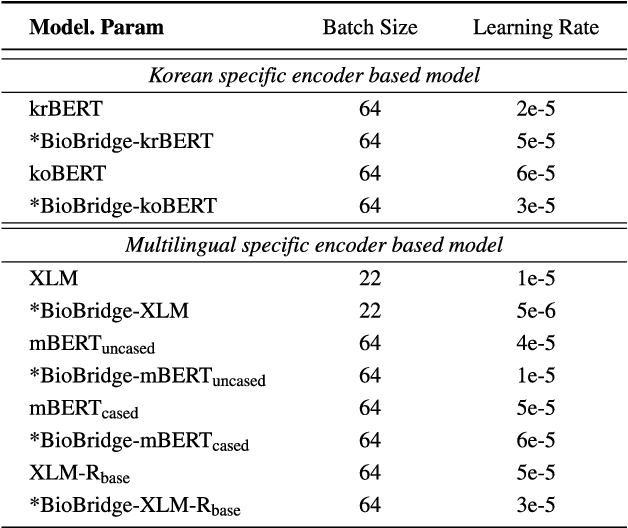
Abstract:Pediatric Emergency Department (PED) overcrowding presents a significant global challenge, prompting the need for efficient solutions. This paper introduces the BioBridge framework, a novel approach that applies Natural Language Processing (NLP) to Electronic Medical Records (EMRs) in written free-text form to enhance decision-making in PED. In non-English speaking countries, such as South Korea, EMR data is often written in a Code-Switching (CS) format that mixes the native language with English, with most code-switched English words having clinical significance. The BioBridge framework consists of two core modules: "bridging modality in context" and "unified bio-embedding." The "bridging modality in context" module improves the contextual understanding of bilingual and code-switched EMRs. In the "unified bio-embedding" module, the knowledge of the model trained in the medical domain is injected into the encoder-based model to bridge the gap between the medical and general domains. Experimental results demonstrate that the proposed BioBridge significantly performance traditional machine learning and pre-trained encoder-based models on several metrics, including F1 score, area under the receiver operating characteristic curve (AUROC), area under the precision-recall curve (AUPRC), and Brier score. Specifically, BioBridge-XLM achieved enhancements of 0.85% in F1 score, 0.75% in AUROC, and 0.76% in AUPRC, along with a notable 3.04% decrease in the Brier score, demonstrating marked improvements in accuracy, reliability, and prediction calibration over the baseline XLM model. The source code will be made publicly available.
* Accepted at IEEE Access 2024
Toward a Well-Calibrated Discrimination via Survival Outcome-Aware Contrastive Learning
Oct 15, 2024Abstract:Previous deep learning approaches for survival analysis have primarily relied on ranking losses to improve discrimination performance, which often comes at the expense of calibration performance. To address such an issue, we propose a novel contrastive learning approach specifically designed to enhance discrimination \textit{without} sacrificing calibration. Our method employs weighted sampling within a contrastive learning framework, assigning lower penalties to samples with similar survival outcomes. This aligns well with the assumption that patients with similar event times share similar clinical statuses. Consequently, when augmented with the commonly used negative log-likelihood loss, our approach significantly improves discrimination performance without directly manipulating the model outputs, thereby achieving better calibration. Experiments on multiple real-world clinical datasets demonstrate that our method outperforms state-of-the-art deep survival models in both discrimination and calibration. Through comprehensive ablation studies, we further validate the effectiveness of our approach through quantitative and qualitative analyses.
Enhancing Anomaly Detection via Generating Diversified and Hard-to-distinguish Synthetic Anomalies
Sep 16, 2024Abstract:Unsupervised anomaly detection is a daunting task, as it relies solely on normality patterns from the training data to identify unseen anomalies during testing. Recent approaches have focused on leveraging domain-specific transformations or perturbations to generate synthetic anomalies from normal samples. The objective here is to acquire insights into normality patterns by learning to differentiate between normal samples and these crafted anomalies. However, these approaches often encounter limitations when domain-specific transformations are not well-specified such as in tabular data, or when it becomes trivial to distinguish between them. To address these issues, we introduce a novel domain-agnostic method that employs a set of conditional perturbators and a discriminator. The perturbators are trained to generate input-dependent perturbations, which are subsequently utilized to construct synthetic anomalies, and the discriminator is trained to distinguish normal samples from them. We ensure that the generated anomalies are both diverse and hard to distinguish through two key strategies: i) directing perturbations to be orthogonal to each other and ii) constraining perturbations to remain in proximity to normal samples. Throughout experiments on real-world datasets, we demonstrate the superiority of our method over state-of-the-art benchmarks, which is evident not only in image data but also in tabular data, where domain-specific transformation is not readily accessible. Additionally, we empirically confirm the adaptability of our method to semi-supervised settings, demonstrating its capacity to incorporate supervised signals to enhance anomaly detection performance even further.
Automatic Quantification of Serial PET/CT Images for Pediatric Hodgkin Lymphoma Patients Using a Longitudinally-Aware Segmentation Network
Apr 12, 2024


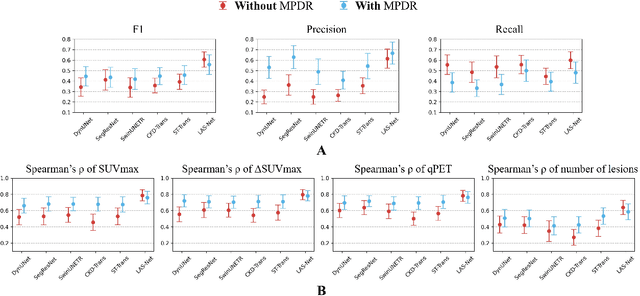
Abstract:$\textbf{Purpose}$: Automatic quantification of longitudinal changes in PET scans for lymphoma patients has proven challenging, as residual disease in interim-therapy scans is often subtle and difficult to detect. Our goal was to develop a longitudinally-aware segmentation network (LAS-Net) that can quantify serial PET/CT images for pediatric Hodgkin lymphoma patients. $\textbf{Materials and Methods}$: This retrospective study included baseline (PET1) and interim (PET2) PET/CT images from 297 patients enrolled in two Children's Oncology Group clinical trials (AHOD1331 and AHOD0831). LAS-Net incorporates longitudinal cross-attention, allowing relevant features from PET1 to inform the analysis of PET2. Model performance was evaluated using Dice coefficients for PET1 and detection F1 scores for PET2. Additionally, we extracted and compared quantitative PET metrics, including metabolic tumor volume (MTV) and total lesion glycolysis (TLG) in PET1, as well as qPET and $\Delta$SUVmax in PET2, against physician measurements. We quantified their agreement using Spearman's $\rho$ correlations and employed bootstrap resampling for statistical analysis. $\textbf{Results}$: LAS-Net detected residual lymphoma in PET2 with an F1 score of 0.606 (precision/recall: 0.615/0.600), outperforming all comparator methods (P<0.01). For baseline segmentation, LAS-Net achieved a mean Dice score of 0.772. In PET quantification, LAS-Net's measurements of qPET, $\Delta$SUVmax, MTV and TLG were strongly correlated with physician measurements, with Spearman's $\rho$ of 0.78, 0.80, 0.93 and 0.96, respectively. The performance remained high, with a slight decrease, in an external testing cohort. $\textbf{Conclusion}$: LAS-Net achieved high performance in quantifying PET metrics across serial scans, highlighting the value of longitudinal awareness in evaluating multi-time-point imaging datasets.
Domain-adapted large language models for classifying nuclear medicine reports
Mar 01, 2023



Abstract:With the growing use of transformer-based language models in medicine, it is unclear how well these models generalize to nuclear medicine which has domain-specific vocabulary and unique reporting styles. In this study, we evaluated the value of domain adaptation in nuclear medicine by adapting language models for the purpose of 5-point Deauville score prediction based on clinical 18F-fluorodeoxyglucose (FDG) PET/CT reports. We retrospectively retrieved 4542 text reports and 1664 images for FDG PET/CT lymphoma exams from 2008-2018 in our clinical imaging database. Deauville scores were removed from the reports and then the remaining text in the reports was used as the model input. Multiple general-purpose transformer language models were used to classify the reports into Deauville scores 1-5. We then adapted the models to the nuclear medicine domain using masked language modeling and assessed its impact on classification performance. The language models were compared against vision models, a multimodal vision language model, and a nuclear medicine physician with seven-fold Monte Carlo cross validation, reported are the mean and standard deviations. Domain adaption improved all language models. For example, BERT improved from 61.3% five-class accuracy to 65.7% following domain adaptation. The best performing model (domain-adapted RoBERTa) achieved a five-class accuracy of 77.4%, which was better than the physician's performance (66%), the best vision model's performance (48.1), and was similar to the multimodal model's performance (77.2). Domain adaptation improved the performance of large language models in interpreting nuclear medicine text reports.
T-Phenotype: Discovering Phenotypes of Predictive Temporal Patterns in Disease Progression
Feb 24, 2023



Abstract:Clustering time-series data in healthcare is crucial for clinical phenotyping to understand patients' disease progression patterns and to design treatment guidelines tailored to homogeneous patient subgroups. While rich temporal dynamics enable the discovery of potential clusters beyond static correlations, two major challenges remain outstanding: i) discovery of predictive patterns from many potential temporal correlations in the multi-variate time-series data and ii) association of individual temporal patterns to the target label distribution that best characterizes the underlying clinical progression. To address such challenges, we develop a novel temporal clustering method, T-Phenotype, to discover phenotypes of predictive temporal patterns from labeled time-series data. We introduce an efficient representation learning approach in frequency domain that can encode variable-length, irregularly-sampled time-series into a unified representation space, which is then applied to identify various temporal patterns that potentially contribute to the target label using a new notion of path-based similarity. Throughout the experiments on synthetic and real-world datasets, we show that T-Phenotype achieves the best phenotype discovery performance over all the evaluated baselines. We further demonstrate the utility of T-Phenotype by uncovering clinically meaningful patient subgroups characterized by unique temporal patterns.
SurvITE: Learning Heterogeneous Treatment Effects from Time-to-Event Data
Oct 26, 2021



Abstract:We study the problem of inferring heterogeneous treatment effects from time-to-event data. While both the related problems of (i) estimating treatment effects for binary or continuous outcomes and (ii) predicting survival outcomes have been well studied in the recent machine learning literature, their combination -- albeit of high practical relevance -- has received considerably less attention. With the ultimate goal of reliably estimating the effects of treatments on instantaneous risk and survival probabilities, we focus on the problem of learning (discrete-time) treatment-specific conditional hazard functions. We find that unique challenges arise in this context due to a variety of covariate shift issues that go beyond a mere combination of well-studied confounding and censoring biases. We theoretically analyse their effects by adapting recent generalization bounds from domain adaptation and treatment effect estimation to our setting and discuss implications for model design. We use the resulting insights to propose a novel deep learning method for treatment-specific hazard estimation based on balancing representations. We investigate performance across a range of experimental settings and empirically confirm that our method outperforms baselines by addressing covariate shifts from various sources.
A Variational Information Bottleneck Approach to Multi-Omics Data Integration
Feb 10, 2021



Abstract:Integration of data from multiple omics techniques is becoming increasingly important in biomedical research. Due to non-uniformity and technical limitations in omics platforms, such integrative analyses on multiple omics, which we refer to as views, involve learning from incomplete observations with various view-missing patterns. This is challenging because i) complex interactions within and across observed views need to be properly addressed for optimal predictive power and ii) observations with various view-missing patterns need to be flexibly integrated. To address such challenges, we propose a deep variational information bottleneck (IB) approach for incomplete multi-view observations. Our method applies the IB framework on marginal and joint representations of the observed views to focus on intra-view and inter-view interactions that are relevant for the target. Most importantly, by modeling the joint representations as a product of marginal representations, we can efficiently learn from observed views with various view-missing patterns. Experiments on real-world datasets show that our method consistently achieves gain from data integration and outperforms state-of-the-art benchmarks.
Temporal Phenotyping using Deep Predictive Clustering of Disease Progression
Jun 15, 2020



Abstract:Due to the wider availability of modern electronic health records, patient care data is often being stored in the form of time-series. Clustering such time-series data is crucial for patient phenotyping, anticipating patients' prognoses by identifying "similar" patients, and designing treatment guidelines that are tailored to homogeneous patient subgroups. In this paper, we develop a deep learning approach for clustering time-series data, where each cluster comprises patients who share similar future outcomes of interest (e.g., adverse events, the onset of comorbidities). To encourage each cluster to have homogeneous future outcomes, the clustering is carried out by learning discrete representations that best describe the future outcome distribution based on novel loss functions. Experiments on two real-world datasets show that our model achieves superior clustering performance over state-of-the-art benchmarks and identifies meaningful clusters that can be translated into actionable information for clinical decision-making.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge