Dongjoon Lee
BioBridge: Unified Bio-Embedding with Bridging Modality in Code-Switched EMR
Dec 16, 2024
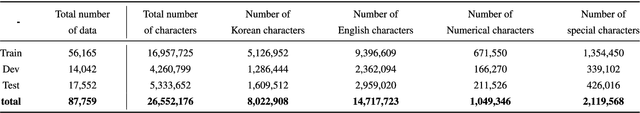
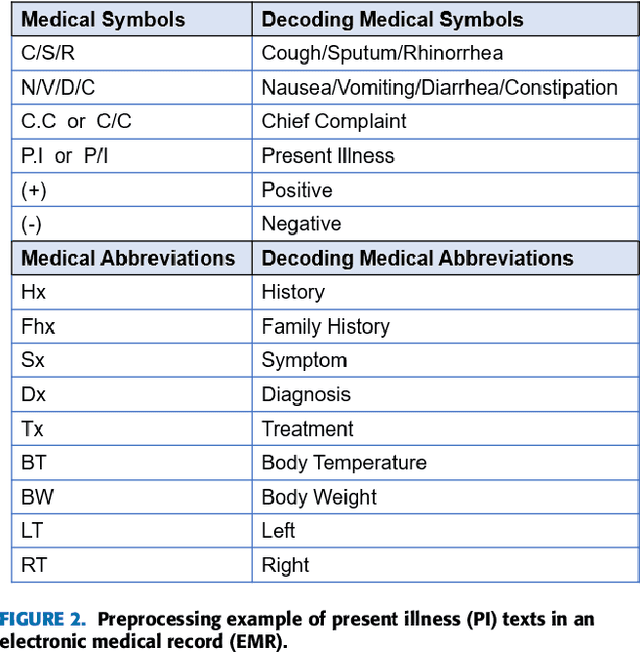
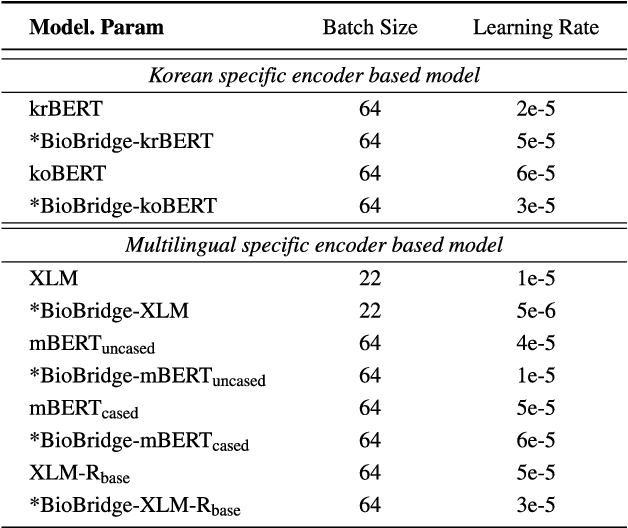
Abstract:Pediatric Emergency Department (PED) overcrowding presents a significant global challenge, prompting the need for efficient solutions. This paper introduces the BioBridge framework, a novel approach that applies Natural Language Processing (NLP) to Electronic Medical Records (EMRs) in written free-text form to enhance decision-making in PED. In non-English speaking countries, such as South Korea, EMR data is often written in a Code-Switching (CS) format that mixes the native language with English, with most code-switched English words having clinical significance. The BioBridge framework consists of two core modules: "bridging modality in context" and "unified bio-embedding." The "bridging modality in context" module improves the contextual understanding of bilingual and code-switched EMRs. In the "unified bio-embedding" module, the knowledge of the model trained in the medical domain is injected into the encoder-based model to bridge the gap between the medical and general domains. Experimental results demonstrate that the proposed BioBridge significantly performance traditional machine learning and pre-trained encoder-based models on several metrics, including F1 score, area under the receiver operating characteristic curve (AUROC), area under the precision-recall curve (AUPRC), and Brier score. Specifically, BioBridge-XLM achieved enhancements of 0.85% in F1 score, 0.75% in AUROC, and 0.76% in AUPRC, along with a notable 3.04% decrease in the Brier score, demonstrating marked improvements in accuracy, reliability, and prediction calibration over the baseline XLM model. The source code will be made publicly available.
* Accepted at IEEE Access 2024
Toward a Well-Calibrated Discrimination via Survival Outcome-Aware Contrastive Learning
Oct 15, 2024Abstract:Previous deep learning approaches for survival analysis have primarily relied on ranking losses to improve discrimination performance, which often comes at the expense of calibration performance. To address such an issue, we propose a novel contrastive learning approach specifically designed to enhance discrimination \textit{without} sacrificing calibration. Our method employs weighted sampling within a contrastive learning framework, assigning lower penalties to samples with similar survival outcomes. This aligns well with the assumption that patients with similar event times share similar clinical statuses. Consequently, when augmented with the commonly used negative log-likelihood loss, our approach significantly improves discrimination performance without directly manipulating the model outputs, thereby achieving better calibration. Experiments on multiple real-world clinical datasets demonstrate that our method outperforms state-of-the-art deep survival models in both discrimination and calibration. Through comprehensive ablation studies, we further validate the effectiveness of our approach through quantitative and qualitative analyses.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge