Xin Tie
Opportunistic Promptable Segmentation: Leveraging Routine Radiological Annotations to Guide 3D CT Lesion Segmentation
Jan 30, 2026Abstract:The development of machine learning models for CT imaging depends on the availability of large, high-quality, and diverse annotated datasets. Although large volumes of CT images and reports are readily available in clinical picture archiving and communication systems (PACS), 3D segmentations of critical findings are costly to obtain, typically requiring extensive manual annotation by radiologists. On the other hand, it is common for radiologists to provide limited annotations of findings during routine reads, such as line measurements and arrows, that are often stored in PACS as GSPS objects. We posit that these sparse annotations can be extracted along with CT volumes and converted into 3D segmentations using promptable segmentation models, a paradigm we term Opportunistic Promptable Segmentation. To enable this paradigm, we propose SAM2CT, the first promptable segmentation model designed to convert radiologist annotations into 3D segmentations in CT volumes. SAM2CT builds upon SAM2 by extending the prompt encoder to support arrow and line inputs and by introducing Memory-Conditioned Memories (MCM), a memory encoding strategy tailored to 3D medical volumes. On public lesion segmentation benchmarks, SAM2CT outperforms existing promptable segmentation models and similarly trained baselines, achieving Dice similarity coefficients of 0.649 for arrow prompts and 0.757 for line prompts. Applying the model to pre-existing GSPS annotations from a clinical PACS (N = 60), SAM2CT generates 3D segmentations that are clinically acceptable or require only minor adjustments in 87% of cases, as scored by radiologists. Additionally, SAM2CT demonstrates strong zero-shot performance on select Emergency Department findings. These results suggest that large-scale mining of historical GSPS annotations represents a promising and scalable approach for generating 3D CT segmentation datasets.
PETAR: Localized Findings Generation with Mask-Aware Vision-Language Modeling for PET Automated Reporting
Oct 31, 2025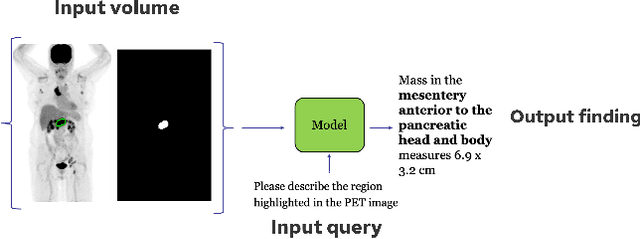
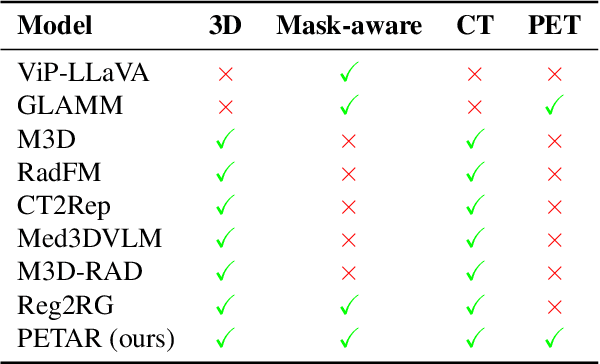
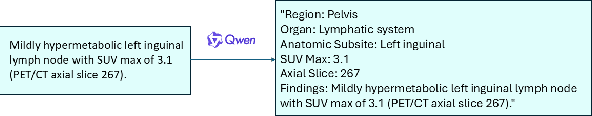
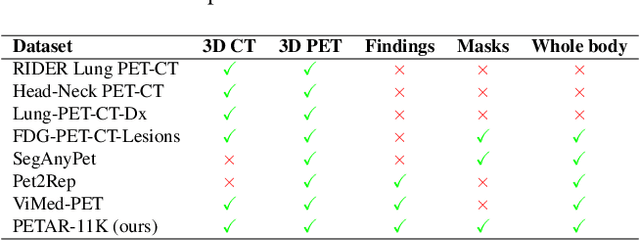
Abstract:Recent advances in vision-language models (VLMs) have enabled impressive multimodal reasoning, yet most medical applications remain limited to 2D imaging. In this work, we extend VLMs to 3D positron emission tomography and computed tomography (PET/CT), a domain characterized by large volumetric data, small and dispersed lesions, and lengthy radiology reports. We introduce a large-scale dataset comprising over 11,000 lesion-level descriptions paired with 3D segmentations from more than 5,000 PET/CT exams, extracted via a hybrid rule-based and large language model (LLM) pipeline. Building upon this dataset, we propose PETAR-4B, a 3D mask-aware vision-language model that integrates PET, CT, and lesion contours for spatially grounded report generation. PETAR bridges global contextual reasoning with fine-grained lesion awareness, producing clinically coherent and localized findings. Comprehensive automated and human evaluations demonstrate that PETAR substantially improves PET/CT report generation quality, advancing 3D medical vision-language understanding.
Deep Learning for Longitudinal Gross Tumor Volume Segmentation in MRI-Guided Adaptive Radiotherapy for Head and Neck Cancer
Dec 01, 2024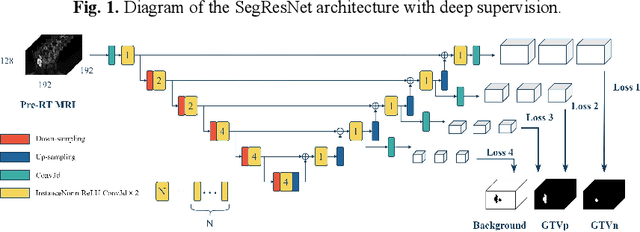
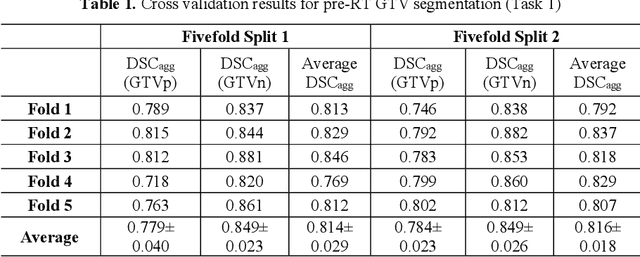
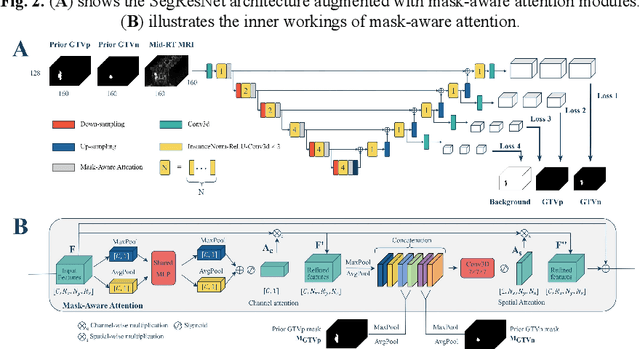
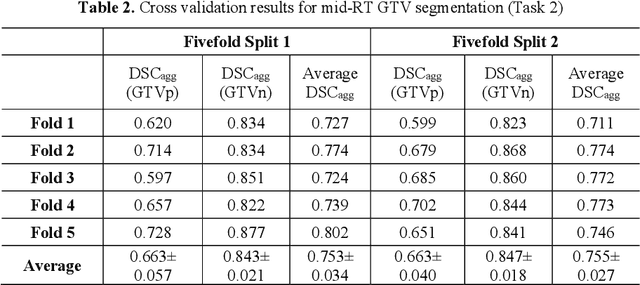
Abstract:Accurate segmentation of gross tumor volume (GTV) is essential for effective MRI-guided adaptive radiotherapy (MRgART) in head and neck cancer. However, manual segmentation of the GTV over the course of therapy is time-consuming and prone to interobserver variability. Deep learning (DL) has the potential to overcome these challenges by automatically delineating GTVs. In this study, our team, $\textit{UW LAIR}$, tackled the challenges of both pre-radiotherapy (pre-RT) (Task 1) and mid-radiotherapy (mid-RT) (Task 2) tumor volume segmentation. To this end, we developed a series of DL models for longitudinal GTV segmentation. The backbone of our models for both tasks was SegResNet with deep supervision. For Task 1, we trained the model using a combined dataset of pre-RT and mid-RT MRI data, which resulted in the improved aggregated Dice similarity coefficient (DSCagg) on an internal testing set compared to models trained solely on pre-RT MRI data. In Task 2, we introduced mask-aware attention modules, enabling pre-RT GTV masks to influence intermediate features learned from mid-RT data. This attention-based approach yielded slight improvements over the baseline method, which concatenated mid-RT MRI with pre-RT GTV masks as input. In the final testing phase, the ensemble of 10 pre-RT segmentation models achieved an average DSCagg of 0.794, with 0.745 for primary GTV (GTVp) and 0.844 for metastatic lymph nodes (GTVn) in Task 1. For Task 2, the ensemble of 10 mid-RT segmentation models attained an average DSCagg of 0.733, with 0.607 for GTVp and 0.859 for GTVn, leading us to $\textbf{achieve 1st place}$. In summary, we presented a collection of DL models that could facilitate GTV segmentation in MRgART, offering the potential to streamline radiation oncology workflows. Our code and model weights are available at https://github.com/xtie97/HNTS-MRG24-UWLAIR.
Automatic Quantification of Serial PET/CT Images for Pediatric Hodgkin Lymphoma Patients Using a Longitudinally-Aware Segmentation Network
Apr 12, 2024


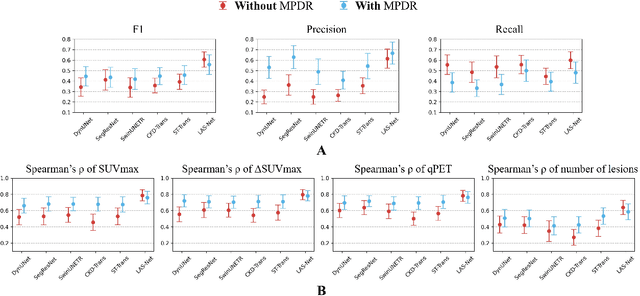
Abstract:$\textbf{Purpose}$: Automatic quantification of longitudinal changes in PET scans for lymphoma patients has proven challenging, as residual disease in interim-therapy scans is often subtle and difficult to detect. Our goal was to develop a longitudinally-aware segmentation network (LAS-Net) that can quantify serial PET/CT images for pediatric Hodgkin lymphoma patients. $\textbf{Materials and Methods}$: This retrospective study included baseline (PET1) and interim (PET2) PET/CT images from 297 patients enrolled in two Children's Oncology Group clinical trials (AHOD1331 and AHOD0831). LAS-Net incorporates longitudinal cross-attention, allowing relevant features from PET1 to inform the analysis of PET2. Model performance was evaluated using Dice coefficients for PET1 and detection F1 scores for PET2. Additionally, we extracted and compared quantitative PET metrics, including metabolic tumor volume (MTV) and total lesion glycolysis (TLG) in PET1, as well as qPET and $\Delta$SUVmax in PET2, against physician measurements. We quantified their agreement using Spearman's $\rho$ correlations and employed bootstrap resampling for statistical analysis. $\textbf{Results}$: LAS-Net detected residual lymphoma in PET2 with an F1 score of 0.606 (precision/recall: 0.615/0.600), outperforming all comparator methods (P<0.01). For baseline segmentation, LAS-Net achieved a mean Dice score of 0.772. In PET quantification, LAS-Net's measurements of qPET, $\Delta$SUVmax, MTV and TLG were strongly correlated with physician measurements, with Spearman's $\rho$ of 0.78, 0.80, 0.93 and 0.96, respectively. The performance remained high, with a slight decrease, in an external testing cohort. $\textbf{Conclusion}$: LAS-Net achieved high performance in quantifying PET metrics across serial scans, highlighting the value of longitudinal awareness in evaluating multi-time-point imaging datasets.
Automatic Personalized Impression Generation for PET Reports Using Large Language Models
Sep 18, 2023Abstract:Purpose: To determine if fine-tuned large language models (LLMs) can generate accurate, personalized impressions for whole-body PET reports. Materials and Methods: Twelve language models were trained on a corpus of PET reports using the teacher-forcing algorithm, with the report findings as input and the clinical impressions as reference. An extra input token encodes the reading physician's identity, allowing models to learn physician-specific reporting styles. Our corpus comprised 37,370 retrospective PET reports collected from our institution between 2010 and 2022. To identify the best LLM, 30 evaluation metrics were benchmarked against quality scores from two nuclear medicine (NM) physicians, with the most aligned metrics selecting the model for expert evaluation. In a subset of data, model-generated impressions and original clinical impressions were assessed by three NM physicians according to 6 quality dimensions and an overall utility score (5-point scale). Each physician reviewed 12 of their own reports and 12 reports from other physicians. Bootstrap resampling was used for statistical analysis. Results: Of all evaluation metrics, domain-adapted BARTScore and PEGASUSScore showed the highest Spearman's rho correlations (0.568 and 0.563) with physician preferences. Based on these metrics, the fine-tuned PEGASUS model was selected as the top LLM. When physicians reviewed PEGASUS-generated impressions in their own style, 89% were considered clinically acceptable, with a mean utility score of 4.08/5. Physicians rated these personalized impressions as comparable in overall utility to the impressions dictated by other physicians (4.03, P=0.41). Conclusion: Personalized impressions generated by PEGASUS were clinically useful, highlighting its potential to expedite PET reporting.
A Generalizable Artificial Intelligence Model for COVID-19 Classification Task Using Chest X-ray Radiographs: Evaluated Over Four Clinical Datasets with 15,097 Patients
Oct 04, 2022



Abstract:Purpose: To answer the long-standing question of whether a model trained from a single clinical site can be generalized to external sites. Materials and Methods: 17,537 chest x-ray radiographs (CXRs) from 3,264 COVID-19-positive patients and 4,802 COVID-19-negative patients were collected from a single site for AI model development. The generalizability of the trained model was retrospectively evaluated using four different real-world clinical datasets with a total of 26,633 CXRs from 15,097 patients (3,277 COVID-19-positive patients). The area under the receiver operating characteristic curve (AUC) was used to assess diagnostic performance. Results: The AI model trained using a single-source clinical dataset achieved an AUC of 0.82 (95% CI: 0.80, 0.84) when applied to the internal temporal test set. When applied to datasets from two external clinical sites, an AUC of 0.81 (95% CI: 0.80, 0.82) and 0.82 (95% CI: 0.80, 0.84) were achieved. An AUC of 0.79 (95% CI: 0.77, 0.81) was achieved when applied to a multi-institutional COVID-19 dataset collected by the Medical Imaging and Data Resource Center (MIDRC). A power-law dependence, N^(k )(k is empirically found to be -0.21 to -0.25), indicates a relatively weak performance dependence on the training data sizes. Conclusion: COVID-19 classification AI model trained using well-curated data from a single clinical site is generalizable to external clinical sites without a significant drop in performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge