Guang-Hong Chen
A Generalizable Artificial Intelligence Model for COVID-19 Classification Task Using Chest X-ray Radiographs: Evaluated Over Four Clinical Datasets with 15,097 Patients
Oct 04, 2022



Abstract:Purpose: To answer the long-standing question of whether a model trained from a single clinical site can be generalized to external sites. Materials and Methods: 17,537 chest x-ray radiographs (CXRs) from 3,264 COVID-19-positive patients and 4,802 COVID-19-negative patients were collected from a single site for AI model development. The generalizability of the trained model was retrospectively evaluated using four different real-world clinical datasets with a total of 26,633 CXRs from 15,097 patients (3,277 COVID-19-positive patients). The area under the receiver operating characteristic curve (AUC) was used to assess diagnostic performance. Results: The AI model trained using a single-source clinical dataset achieved an AUC of 0.82 (95% CI: 0.80, 0.84) when applied to the internal temporal test set. When applied to datasets from two external clinical sites, an AUC of 0.81 (95% CI: 0.80, 0.82) and 0.82 (95% CI: 0.80, 0.84) were achieved. An AUC of 0.79 (95% CI: 0.77, 0.81) was achieved when applied to a multi-institutional COVID-19 dataset collected by the Medical Imaging and Data Resource Center (MIDRC). A power-law dependence, N^(k )(k is empirically found to be -0.21 to -0.25), indicates a relatively weak performance dependence on the training data sizes. Conclusion: COVID-19 classification AI model trained using well-curated data from a single clinical site is generalizable to external clinical sites without a significant drop in performance.
Deep Learning Angiography (DLA): Three-dimensional C-arm Cone Beam CT Angiography Using Deep Learning
Jan 26, 2018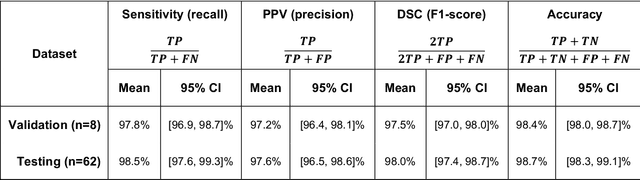
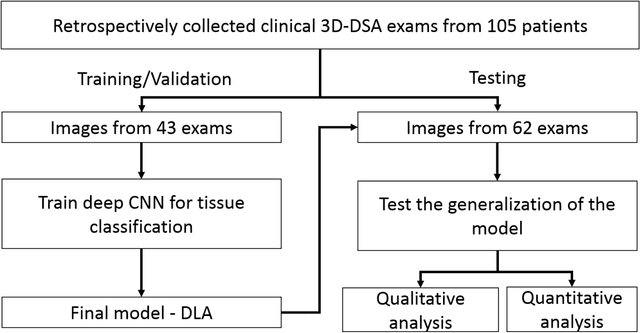
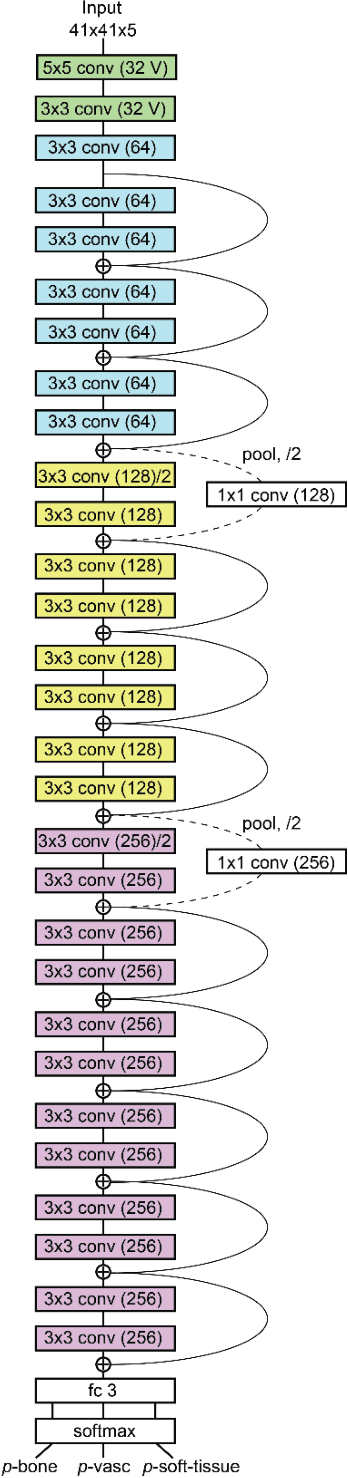
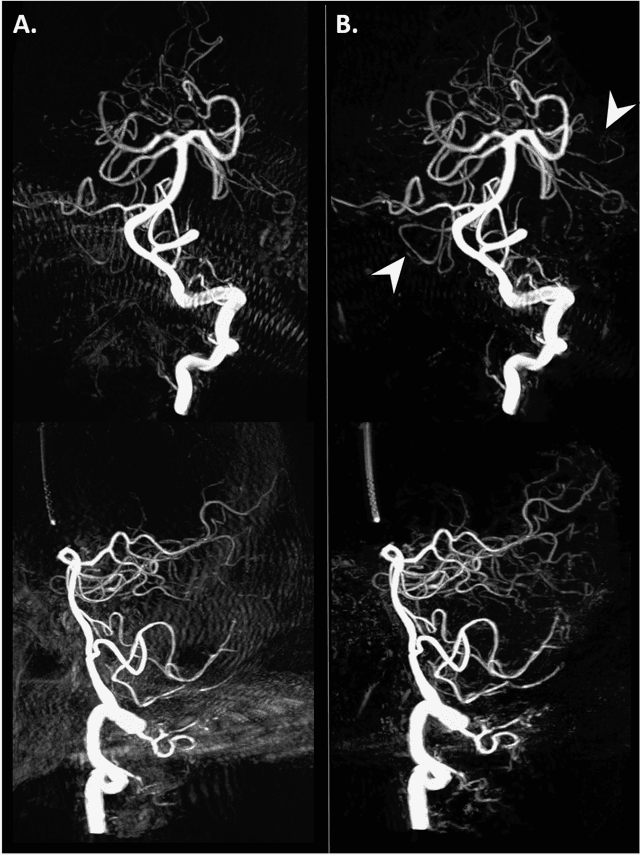
Abstract:Background and Purpose: Our purpose was to develop a deep learning angiography (DLA) method to generate 3D cerebral angiograms from a single contrast-enhanced acquisition. Material and Methods: Under an approved IRB protocol 105 3D-DSA exams were randomly selected from an internal database. All were acquired using a clinical system (Axiom Artis zee, Siemens Healthineers) in conjunction with a standard injection protocol. More than 150 million labeled voxels from 35 subjects were used for training. A deep convolutional neural network was trained to classify each image voxel into three tissue types (vasculature, bone and soft tissue). The trained DLA model was then applied for tissue classification in a validation cohort of 8 subjects and a final testing cohort consisting of the remaining 62 subjects. The final vasculature tissue class was used to generate the 3D-DLA images. To quantify the generalization error of the trained model, accuracy, sensitivity, precision and F1-scores were calculated for vasculature classification in relevant anatomy. The 3D-DLA and clinical 3D-DSA images were subject to a qualitative assessment for the presence of inter-sweep motion artifacts. Results: Vasculature classification accuracy and 95% CI in the testing dataset was 98.7% ([98.3, 99.1] %). No residual signal from osseous structures was observed for all 3D-DLA testing cases except for small regions in the otic capsule and nasal cavity compared to 37% (23/62) of the 3D-DSAs. Conclusion: DLA accurately recreated the vascular anatomy of the 3D-DSA reconstructions without mask. DLA reduced mis-registration artifacts induced by inter-sweep motion. DLA reduces radiation exposure required to obtain clinically useful 3D-DSA
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge