Javier Alvarez-Valle
NOVA: An Agentic Framework for Automated Histopathology Analysis and Discovery
Nov 14, 2025



Abstract:Digitized histopathology analysis involves complex, time-intensive workflows and specialized expertise, limiting its accessibility. We introduce NOVA, an agentic framework that translates scientific queries into executable analysis pipelines by iteratively generating and running Python code. NOVA integrates 49 domain-specific tools (e.g., nuclei segmentation, whole-slide encoding) built on open-source software, and can also create new tools ad hoc. To evaluate such systems, we present SlideQuest, a 90-question benchmark -- verified by pathologists and biomedical scientists -- spanning data processing, quantitative analysis, and hypothesis testing. Unlike prior biomedical benchmarks focused on knowledge recall or diagnostic QA, SlideQuest demands multi-step reasoning, iterative coding, and computational problem solving. Quantitative evaluation shows NOVA outperforms coding-agent baselines, and a pathologist-verified case study links morphology to prognostically relevant PAM50 subtypes, demonstrating its scalable discovery potential.
Data Scaling Laws for Radiology Foundation Models
Sep 16, 2025Abstract:Foundation vision encoders such as CLIP and DINOv2, trained on web-scale data, exhibit strong transfer performance across tasks and datasets. However, medical imaging foundation models remain constrained by smaller datasets, limiting our understanding of how data scale and pretraining paradigms affect performance in this setting. In this work, we systematically study continual pretraining of two vision encoders, MedImageInsight (MI2) and RAD-DINO representing the two major encoder paradigms CLIP and DINOv2, on up to 3.5M chest x-rays from a single institution, holding compute and evaluation protocols constant. We evaluate on classification (radiology findings, lines and tubes), segmentation (lines and tubes), and radiology report generation. While prior work has primarily focused on tasks related to radiology findings, we include lines and tubes tasks to counterbalance this bias and evaluate a model's ability to extract features that preserve continuity along elongated structures. Our experiments show that MI2 scales more effectively for finding-related tasks, while RAD-DINO is stronger on tube-related tasks. Surprisingly, continually pretraining MI2 with both reports and structured labels using UniCL improves performance, underscoring the value of structured supervision at scale. We further show that for some tasks, as few as 30k in-domain samples are sufficient to surpass open-weights foundation models. These results highlight the utility of center-specific continual pretraining, enabling medical institutions to derive significant performance gains by utilizing in-domain data.
Lunguage: A Benchmark for Structured and Sequential Chest X-ray Interpretation
May 27, 2025Abstract:Radiology reports convey detailed clinical observations and capture diagnostic reasoning that evolves over time. However, existing evaluation methods are limited to single-report settings and rely on coarse metrics that fail to capture fine-grained clinical semantics and temporal dependencies. We introduce LUNGUAGE,a benchmark dataset for structured radiology report generation that supports both single-report evaluation and longitudinal patient-level assessment across multiple studies. It contains 1,473 annotated chest X-ray reports, each reviewed by experts, and 80 of them contain longitudinal annotations to capture disease progression and inter-study intervals, also reviewed by experts. Using this benchmark, we develop a two-stage framework that transforms generated reports into fine-grained, schema-aligned structured representations, enabling longitudinal interpretation. We also propose LUNGUAGESCORE, an interpretable metric that compares structured outputs at the entity, relation, and attribute level while modeling temporal consistency across patient timelines. These contributions establish the first benchmark dataset, structuring framework, and evaluation metric for sequential radiology reporting, with empirical results demonstrating that LUNGUAGESCORE effectively supports structured report evaluation. The code is available at: https://github.com/SuperSupermoon/Lunguage
MAIRA-Seg: Enhancing Radiology Report Generation with Segmentation-Aware Multimodal Large Language Models
Nov 18, 2024



Abstract:There is growing interest in applying AI to radiology report generation, particularly for chest X-rays (CXRs). This paper investigates whether incorporating pixel-level information through segmentation masks can improve fine-grained image interpretation of multimodal large language models (MLLMs) for radiology report generation. We introduce MAIRA-Seg, a segmentation-aware MLLM framework designed to utilize semantic segmentation masks alongside CXRs for generating radiology reports. We train expert segmentation models to obtain mask pseudolabels for radiology-specific structures in CXRs. Subsequently, building on the architectures of MAIRA, a CXR-specialised model for report generation, we integrate a trainable segmentation tokens extractor that leverages these mask pseudolabels, and employ mask-aware prompting to generate draft radiology reports. Our experiments on the publicly available MIMIC-CXR dataset show that MAIRA-Seg outperforms non-segmentation baselines. We also investigate set-of-marks prompting with MAIRA and find that MAIRA-Seg consistently demonstrates comparable or superior performance. The results confirm that using segmentation masks enhances the nuanced reasoning of MLLMs, potentially contributing to better clinical outcomes.
PadChest-GR: A Bilingual Chest X-ray Dataset for Grounded Radiology Report Generation
Nov 07, 2024



Abstract:Radiology report generation (RRG) aims to create free-text radiology reports from clinical imaging. Grounded radiology report generation (GRRG) extends RRG by including the localisation of individual findings on the image. Currently, there are no manually annotated chest X-ray (CXR) datasets to train GRRG models. In this work, we present a dataset called PadChest-GR (Grounded-Reporting) derived from PadChest aimed at training GRRG models for CXR images. We curate a public bi-lingual dataset of 4,555 CXR studies with grounded reports (3,099 abnormal and 1,456 normal), each containing complete lists of sentences describing individual present (positive) and absent (negative) findings in English and Spanish. In total, PadChest-GR contains 7,037 positive and 3,422 negative finding sentences. Every positive finding sentence is associated with up to two independent sets of bounding boxes labelled by different readers and has categorical labels for finding type, locations, and progression. To the best of our knowledge, PadChest-GR is the first manually curated dataset designed to train GRRG models for understanding and interpreting radiological images and generated text. By including detailed localization and comprehensive annotations of all clinically relevant findings, it provides a valuable resource for developing and evaluating GRRG models from CXR images. PadChest-GR can be downloaded under request from https://bimcv.cipf.es/bimcv-projects/padchest-gr/
MedImageInsight: An Open-Source Embedding Model for General Domain Medical Imaging
Oct 09, 2024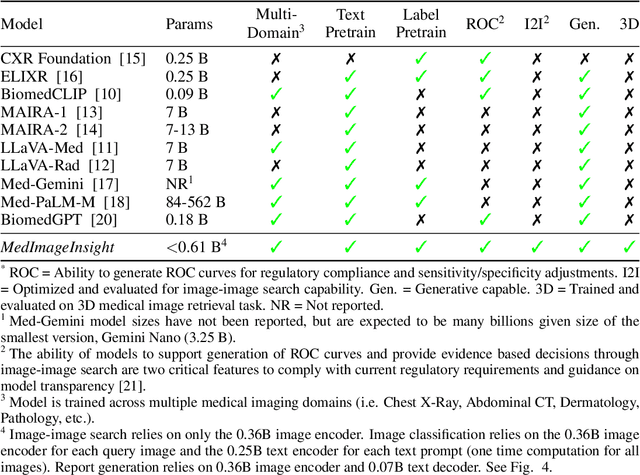
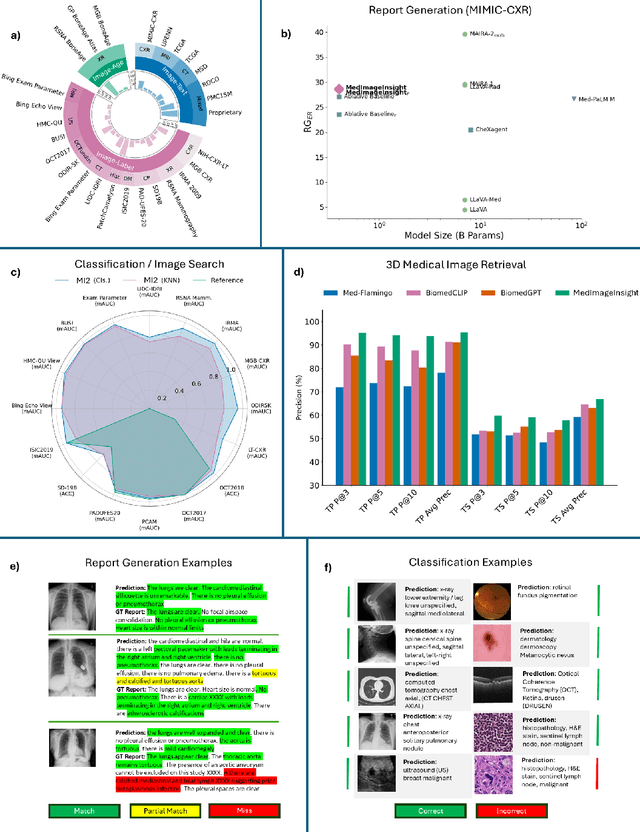
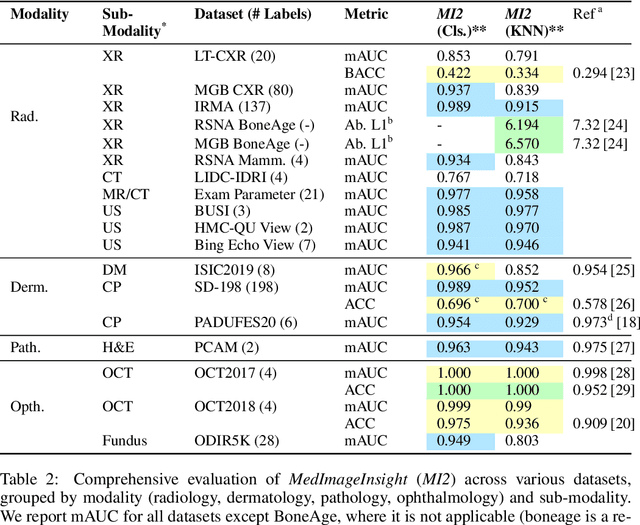
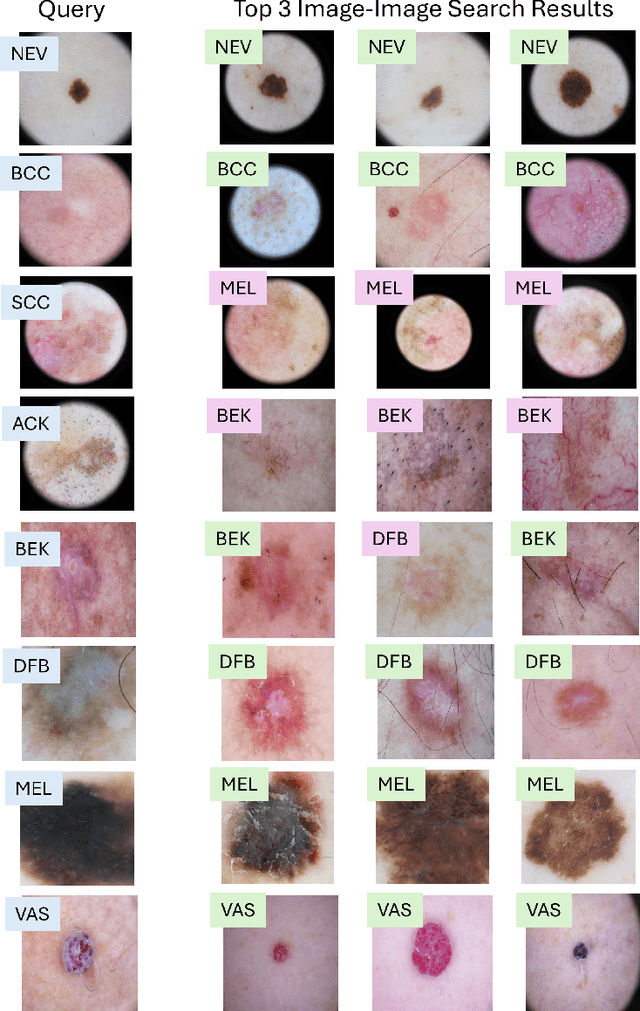
Abstract:In this work, we present MedImageInsight, an open-source medical imaging embedding model. MedImageInsight is trained on medical images with associated text and labels across a diverse collection of domains, including X-Ray, CT, MRI, dermoscopy, OCT, fundus photography, ultrasound, histopathology, and mammography. Rigorous evaluations demonstrate MedImageInsight's ability to achieve state-of-the-art (SOTA) or human expert level performance across classification, image-image search, and fine-tuning tasks. Specifically, on public datasets, MedImageInsight achieves SOTA in CT 3D medical image retrieval, as well as SOTA in disease classification and search for chest X-ray, dermatology, and OCT imaging. Furthermore, MedImageInsight achieves human expert performance in bone age estimation (on both public and partner data), as well as AUC above 0.9 in most other domains. When paired with a text decoder, MedImageInsight achieves near SOTA level single image report findings generation with less than 10\% the parameters of other models. Compared to fine-tuning GPT-4o with only MIMIC-CXR data for the same task, MedImageInsight outperforms in clinical metrics, but underperforms on lexical metrics where GPT-4o sets a new SOTA. Importantly for regulatory purposes, MedImageInsight can generate ROC curves, adjust sensitivity and specificity based on clinical need, and provide evidence-based decision support through image-image search (which can also enable retrieval augmented generation). In an independent clinical evaluation of image-image search in chest X-ray, MedImageInsight outperformed every other publicly available foundation model evaluated by large margins (over 6 points AUC), and significantly outperformed other models in terms of AI fairness (across age and gender). We hope releasing MedImageInsight will help enhance collective progress in medical imaging AI research and development.
MAIRA-2: Grounded Radiology Report Generation
Jun 06, 2024



Abstract:Radiology reporting is a complex task that requires detailed image understanding, integration of multiple inputs, including comparison with prior imaging, and precise language generation. This makes it ideal for the development and use of generative multimodal models. Here, we extend report generation to include the localisation of individual findings on the image - a task we call grounded report generation. Prior work indicates that grounding is important for clarifying image understanding and interpreting AI-generated text. Therefore, grounded reporting stands to improve the utility and transparency of automated report drafting. To enable evaluation of grounded reporting, we propose a novel evaluation framework - RadFact - leveraging the reasoning capabilities of large language models (LLMs). RadFact assesses the factuality of individual generated sentences, as well as correctness of generated spatial localisations when present. We introduce MAIRA-2, a large multimodal model combining a radiology-specific image encoder with a LLM, and trained for the new task of grounded report generation on chest X-rays. MAIRA-2 uses more comprehensive inputs than explored previously: the current frontal image, the current lateral image, the prior frontal image and prior report, as well as the Indication, Technique and Comparison sections of the current report. We demonstrate that these additions significantly improve report quality and reduce hallucinations, establishing a new state of the art on findings generation (without grounding) on MIMIC-CXR while demonstrating the feasibility of grounded reporting as a novel and richer task.
Challenges for Responsible AI Design and Workflow Integration in Healthcare: A Case Study of Automatic Feeding Tube Qualification in Radiology
May 08, 2024
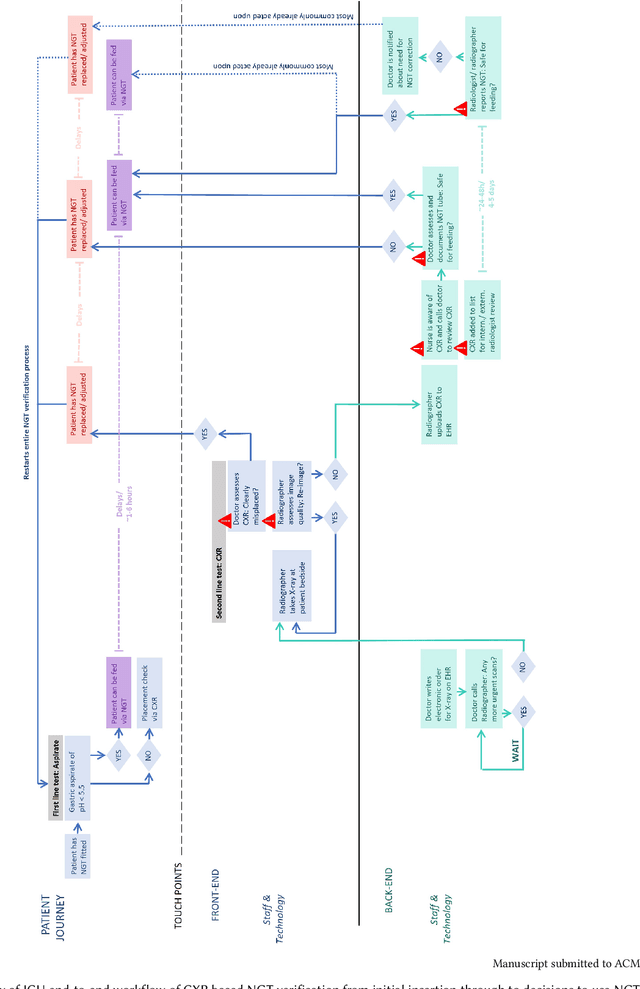

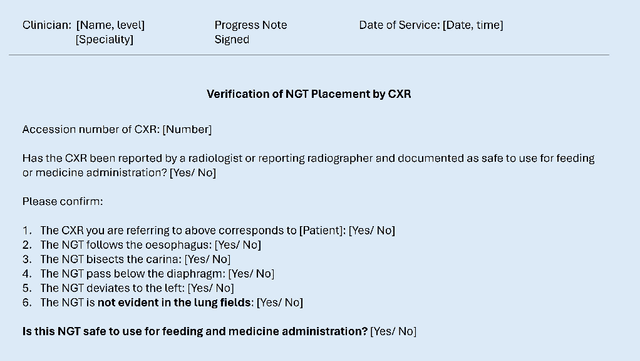
Abstract:Nasogastric tubes (NGTs) are feeding tubes that are inserted through the nose into the stomach to deliver nutrition or medication. If not placed correctly, they can cause serious harm, even death to patients. Recent AI developments demonstrate the feasibility of robustly detecting NGT placement from Chest X-ray images to reduce risks of sub-optimally or critically placed NGTs being missed or delayed in their detection, but gaps remain in clinical practice integration. In this study, we present a human-centered approach to the problem and describe insights derived following contextual inquiry and in-depth interviews with 15 clinical stakeholders. The interviews helped understand challenges in existing workflows, and how best to align technical capabilities with user needs and expectations. We discovered the trade-offs and complexities that need consideration when choosing suitable workflow stages, target users, and design configurations for different AI proposals. We explored how to balance AI benefits and risks for healthcare staff and patients within broader organizational and medical-legal constraints. We also identified data issues related to edge cases and data biases that affect model training and evaluation; how data documentation practices influence data preparation and labelling; and how to measure relevant AI outcomes reliably in future evaluations. We discuss how our work informs design and development of AI applications that are clinically useful, ethical, and acceptable in real-world healthcare services.
RAD-DINO: Exploring Scalable Medical Image Encoders Beyond Text Supervision
Jan 19, 2024Abstract:Language-supervised pre-training has proven to be a valuable method for extracting semantically meaningful features from images, serving as a foundational element in multimodal systems within the computer vision and medical imaging domains. However, resulting features are limited by the information contained within the text. This is particularly problematic in medical imaging, where radiologists' written findings focus on specific observations; a challenge compounded by the scarcity of paired imaging-text data due to concerns over leakage of personal health information. In this work, we fundamentally challenge the prevailing reliance on language supervision for learning general purpose biomedical imaging encoders. We introduce RAD-DINO, a biomedical image encoder pre-trained solely on unimodal biomedical imaging data that obtains similar or greater performance than state-of-the-art biomedical language supervised models on a diverse range of benchmarks. Specifically, the quality of learned representations is evaluated on standard imaging tasks (classification and semantic segmentation), and a vision-language alignment task (text report generation from images). To further demonstrate the drawback of language supervision, we show that features from RAD-DINO correlate with other medical records (e.g., sex or age) better than language-supervised models, which are generally not mentioned in radiology reports. Finally, we conduct a series of ablations determining the factors in RAD-DINO's performance; notably, we observe that RAD-DINO's downstream performance scales well with the quantity and diversity of training data, demonstrating that image-only supervision is a scalable approach for training a foundational biomedical image encoder.
RadEdit: stress-testing biomedical vision models via diffusion image editing
Dec 21, 2023Abstract:Biomedical imaging datasets are often small and biased, meaning that real-world performance of predictive models can be substantially lower than expected from internal testing. This work proposes using generative image editing to simulate dataset shifts and diagnose failure modes of biomedical vision models; this can be used in advance of deployment to assess readiness, potentially reducing cost and patient harm. Existing editing methods can produce undesirable changes, with spurious correlations learned due to the co-occurrence of disease and treatment interventions, limiting practical applicability. To address this, we train a text-to-image diffusion model on multiple chest X-ray datasets and introduce a new editing method RadEdit that uses multiple masks, if present, to constrain changes and ensure consistency in the edited images. We consider three types of dataset shifts: acquisition shift, manifestation shift, and population shift, and demonstrate that our approach can diagnose failures and quantify model robustness without additional data collection, complementing more qualitative tools for explainable AI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge