Lars Heiliger
ReXamine-Global: A Framework for Uncovering Inconsistencies in Radiology Report Generation Metrics
Aug 29, 2024Abstract:Given the rapidly expanding capabilities of generative AI models for radiology, there is a need for robust metrics that can accurately measure the quality of AI-generated radiology reports across diverse hospitals. We develop ReXamine-Global, a LLM-powered, multi-site framework that tests metrics across different writing styles and patient populations, exposing gaps in their generalization. First, our method tests whether a metric is undesirably sensitive to reporting style, providing different scores depending on whether AI-generated reports are stylistically similar to ground-truth reports or not. Second, our method measures whether a metric reliably agrees with experts, or whether metric and expert scores of AI-generated report quality diverge for some sites. Using 240 reports from 6 hospitals around the world, we apply ReXamine-Global to 7 established report evaluation metrics and uncover serious gaps in their generalizability. Developers can apply ReXamine-Global when designing new report evaluation metrics, ensuring their robustness across sites. Additionally, our analysis of existing metrics can guide users of those metrics towards evaluation procedures that work reliably at their sites of interest.
On the Impact of Cross-Domain Data on German Language Models
Oct 13, 2023Abstract:Traditionally, large language models have been either trained on general web crawls or domain-specific data. However, recent successes of generative large language models, have shed light on the benefits of cross-domain datasets. To examine the significance of prioritizing data diversity over quality, we present a German dataset comprising texts from five domains, along with another dataset aimed at containing high-quality data. Through training a series of models ranging between 122M and 750M parameters on both datasets, we conduct a comprehensive benchmark on multiple downstream tasks. Our findings demonstrate that the models trained on the cross-domain dataset outperform those trained on quality data alone, leading to improvements up to $4.45\%$ over the previous state-of-the-art. The models are available at https://huggingface.co/ikim-uk-essen
Multimodal Interactive Lung Lesion Segmentation: A Framework for Annotating PET/CT Images based on Physiological and Anatomical Cues
Jan 24, 2023



Abstract:Recently, deep learning enabled the accurate segmentation of various diseases in medical imaging. These performances, however, typically demand large amounts of manual voxel annotations. This tedious process for volumetric data becomes more complex when not all required information is available in a single imaging domain as is the case for PET/CT data. We propose a multimodal interactive segmentation framework that mitigates these issues by combining anatomical and physiological cues from PET/CT data. Our framework utilizes the geodesic distance transform to represent the user annotations and we implement a novel ellipsoid-based user simulation scheme during training. We further propose two annotation interfaces and conduct a user study to estimate their usability. We evaluated our model on the in-domain validation dataset and an unseen PET/CT dataset. We make our code publicly available: https://github.com/verena-hallitschke/pet-ct-annotate.
AutoPET Challenge: Combining nn-Unet with Swin UNETR Augmented by Maximum Intensity Projection Classifier
Sep 02, 2022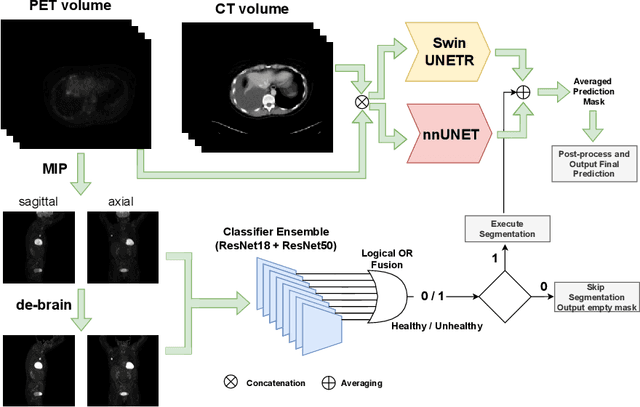
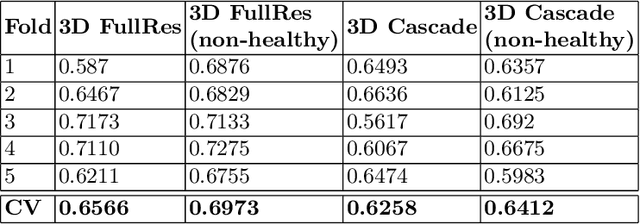
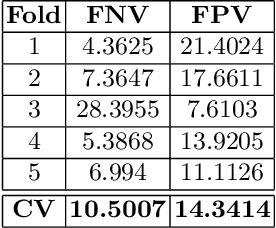
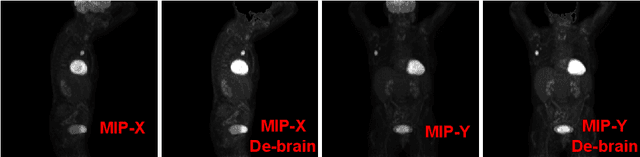
Abstract:Tumor volume and changes in tumor characteristics over time are important biomarkers for cancer therapy. In this context, FDG-PET/CT scans are routinely used for staging and re-staging of cancer, as the radiolabeled fluorodeoxyglucose is taken up in regions of high metabolism. Unfortunately, these regions with high metabolism are not specific to tumors and can also represent physiological uptake by normal functioning organs, inflammation, or infection, making detailed and reliable tumor segmentation in these scans a demanding task. This gap in research is addressed by the AutoPET challenge, which provides a public data set with FDG-PET/CT scans from 900 patients to encourage further improvement in this field. Our contribution to this challenge is an ensemble of two state-of-the-art segmentation models, the nn-Unet and the Swin UNETR, augmented by a maximum intensity projection classifier that acts like a gating mechanism. If it predicts the existence of lesions, both segmentations are combined by a late fusion approach. Our solution achieves a Dice score of 72.12\% on patients diagnosed with lung cancer, melanoma, and lymphoma in our cross-validation. Code: https://github.com/heiligerl/autopet_submission
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge