Fongci Lin
CDEMapper: Enhancing NIH Common Data Element Normalization using Large Language Models
Nov 30, 2024Abstract:Common Data Elements (CDEs) standardize data collection and sharing across studies, enhancing data interoperability and improving research reproducibility. However, implementing CDEs presents challenges due to the broad range and variety of data elements. This study aims to develop an effective and efficient mapping tool to bridge the gap between local data elements and National Institutes of Health (NIH) CDEs. We propose CDEMapper, a large language model (LLM) powered mapping tool designed to assist in mapping local data elements to NIH CDEs. CDEMapper has three core modules: (1) CDE indexing and embeddings. NIH CDEs were indexed and embedded to support semantic search; (2) CDE recommendations. The tool combines Elasticsearch (BM25 similarity methods) with state of the art GPT services to recommend candidate CDEs and their permissible values; and (3) Human review. Users review and select the NIH CDEs and values that best match their data elements and value sets. We evaluate the tool recommendation accuracy against manually annotated mapping results. CDEMapper offers a publicly available, LLM-powered, and intuitive user interface that consolidates essential and advanced mapping services into a streamlined pipeline. It provides a step by step, quality assured mapping workflow designed with a user-centered approach. The evaluation results demonstrated that augmenting BM25 with GPT embeddings and a ranker consistently enhances CDEMapper mapping accuracy in three different mapping settings across four evaluation datasets. This work opens up the potential of using LLMs to assist with CDE recommendation and human curation when aligning local data elements with NIH CDEs. Additionally, this effort enhances clinical research data interoperability and helps researchers better understand the gaps between local data elements and NIH CDEs.
Me LLaMA: Foundation Large Language Models for Medical Applications
Feb 20, 2024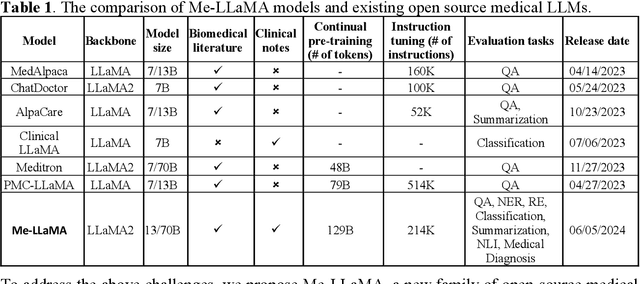



Abstract:Recent large language models (LLMs) like ChatGPT and LLaMA have shown great promise in many AI applications. However, their performance on medical tasks is suboptimal and can be further improved by training on large domain-specific datasets. This study introduces Me LLaMA, a medical LLM family including foundation models - Me LLaMA 13/70B and their chat-enhanced versions - Me LLaMA 13/70B-chat, developed through the continual pre-training and instruction tuning of LLaMA2 using large medical data. Our domain-specific data suite for training and evaluation, includes a large-scale continual pre-training dataset with 129B tokens, an instruction tuning dataset with 214k samples, and a medical evaluation benchmark (MIBE) across six tasks with 14 datasets. Our extensive evaluation using MIBE shows that Me LLaMA models surpass existing open-source medical LLMs in zero-shot and few-shot learning and outperform commercial giants like ChatGPT on 6 out of 8 datasets and GPT-4 in 3 out of 8 datasets. In addition, we empirically investigated the catastrophic forgetting problem, and our results show that Me LLaMA models outperform other medical LLMs. Me LLaMA is one of the first and largest open-source foundational LLMs designed for the medical domain, using both biomedical and clinical data. It exhibits superior performance across both general and medical tasks compared to other medical LLMs, rendering it an attractive choice for medical AI applications. All resources are available at: https://github.com/BIDS-Xu-Lab/Me-LLaMA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge