Ruey-Ling Weng
EHRNavigator: A Multi-Agent System for Patient-Level Clinical Question Answering over Heterogeneous Electronic Health Records
Jan 15, 2026Abstract:Clinical decision-making increasingly relies on timely and context-aware access to patient information within Electronic Health Records (EHRs), yet most existing natural language question-answering (QA) systems are evaluated solely on benchmark datasets, limiting their practical relevance. To overcome this limitation, we introduce EHRNavigator, a multi-agent framework that harnesses AI agents to perform patient-level question answering across heterogeneous and multimodal EHR data. We assessed its performance using both public benchmark and institutional datasets under realistic hospital conditions characterized by diverse schemas, temporal reasoning demands, and multimodal evidence integration. Through quantitative evaluation and clinician-validated chart review, EHRNavigator demonstrated strong generalization, achieving 86% accuracy on real-world cases while maintaining clinically acceptable response times. Overall, these findings confirm that EHRNavigator effectively bridges the gap between benchmark evaluation and clinical deployment, offering a robust, adaptive, and efficient solution for real-world EHR question answering.
Enhancing Financial Time-Series Forecasting with Retrieval-Augmented Large Language Models
Feb 11, 2025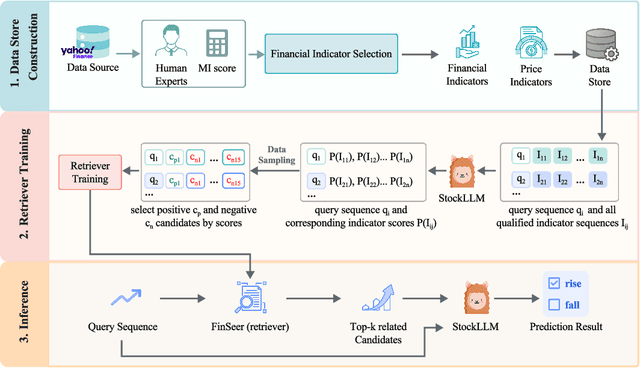
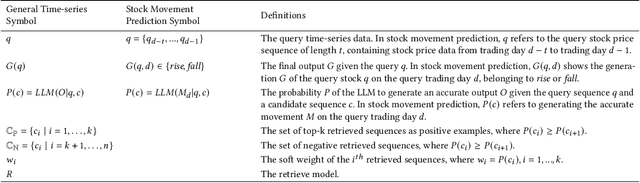
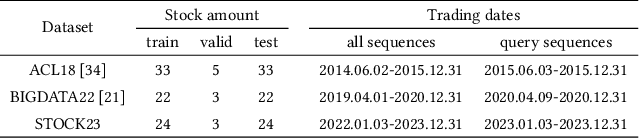
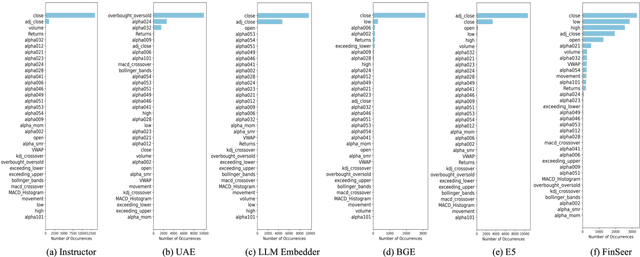
Abstract:Stock movement prediction, a critical task in financial time-series forecasting, relies on identifying and retrieving key influencing factors from vast and complex datasets. However, traditional text-trained or numeric similarity-based retrieval methods often struggle to handle the intricacies of financial data. To address this, we propose the first retrieval-augmented generation (RAG) framework specifically designed for financial time-series forecasting. Our framework incorporates three key innovations: a fine-tuned 1B large language model (StockLLM) as its backbone, a novel candidate selection method enhanced by LLM feedback, and a training objective that maximizes the similarity between queries and historically significant sequences. These advancements enable our retriever, FinSeer, to uncover meaningful patterns while effectively minimizing noise in complex financial datasets. To support robust evaluation, we also construct new datasets that integrate financial indicators and historical stock prices. Experimental results demonstrate that our RAG framework outperforms both the baseline StockLLM and random retrieval methods, showcasing its effectiveness. FinSeer, as the retriever, achieves an 8% higher accuracy on the BIGDATA22 benchmark and retrieves more impactful sequences compared to existing retrieval methods. This work highlights the importance of tailored retrieval models in financial forecasting and provides a novel, scalable framework for future research in the field.
Retrieval-augmented Large Language Models for Financial Time Series Forecasting
Feb 09, 2025



Abstract:Stock movement prediction, a fundamental task in financial time-series forecasting, requires identifying and retrieving critical influencing factors from vast amounts of time-series data. However, existing text-trained or numeric similarity-based retrieval methods fall short in handling complex financial analysis. To address this, we propose the first retrieval-augmented generation (RAG) framework for financial time-series forecasting, featuring three key innovations: a fine-tuned 1B parameter large language model (StockLLM) as the backbone, a novel candidate selection method leveraging LLM feedback, and a training objective that maximizes similarity between queries and historically significant sequences. This enables our retriever, FinSeer, to uncover meaningful patterns while minimizing noise in complex financial data. We also construct new datasets integrating financial indicators and historical stock prices to train FinSeer and ensure robust evaluation. Experimental results demonstrate that our RAG framework outperforms bare StockLLM and random retrieval, highlighting its effectiveness, while FinSeer surpasses existing retrieval methods, achieving an 8\% higher accuracy on BIGDATA22 and retrieving more impactful sequences. This work underscores the importance of tailored retrieval models in financial forecasting and provides a novel framework for future research.
CDEMapper: Enhancing NIH Common Data Element Normalization using Large Language Models
Nov 30, 2024Abstract:Common Data Elements (CDEs) standardize data collection and sharing across studies, enhancing data interoperability and improving research reproducibility. However, implementing CDEs presents challenges due to the broad range and variety of data elements. This study aims to develop an effective and efficient mapping tool to bridge the gap between local data elements and National Institutes of Health (NIH) CDEs. We propose CDEMapper, a large language model (LLM) powered mapping tool designed to assist in mapping local data elements to NIH CDEs. CDEMapper has three core modules: (1) CDE indexing and embeddings. NIH CDEs were indexed and embedded to support semantic search; (2) CDE recommendations. The tool combines Elasticsearch (BM25 similarity methods) with state of the art GPT services to recommend candidate CDEs and their permissible values; and (3) Human review. Users review and select the NIH CDEs and values that best match their data elements and value sets. We evaluate the tool recommendation accuracy against manually annotated mapping results. CDEMapper offers a publicly available, LLM-powered, and intuitive user interface that consolidates essential and advanced mapping services into a streamlined pipeline. It provides a step by step, quality assured mapping workflow designed with a user-centered approach. The evaluation results demonstrated that augmenting BM25 with GPT embeddings and a ranker consistently enhances CDEMapper mapping accuracy in three different mapping settings across four evaluation datasets. This work opens up the potential of using LLMs to assist with CDE recommendation and human curation when aligning local data elements with NIH CDEs. Additionally, this effort enhances clinical research data interoperability and helps researchers better understand the gaps between local data elements and NIH CDEs.
Information Extraction from Clinical Notes: Are We Ready to Switch to Large Language Models?
Nov 15, 2024



Abstract:Backgrounds: Information extraction (IE) is critical in clinical natural language processing (NLP). While large language models (LLMs) excel on generative tasks, their performance on extractive tasks remains debated. Methods: We investigated Named Entity Recognition (NER) and Relation Extraction (RE) using 1,588 clinical notes from four sources (UT Physicians, MTSamples, MIMIC-III, and i2b2). We developed an annotated corpus covering 4 clinical entities and 16 modifiers, and compared instruction-tuned LLaMA-2 and LLaMA-3 against BiomedBERT in terms of performance, generalizability, computational resources, and throughput to BiomedBERT. Results: LLaMA models outperformed BiomedBERT across datasets. With sufficient training data, LLaMA showed modest improvements (1% on NER, 1.5-3.7% on RE); improvements were larger with limited training data. On unseen i2b2 data, LLaMA-3-70B outperformed BiomedBERT by 7% (F1) on NER and 4% on RE. However, LLaMA models required more computing resources and ran up to 28 times slower. We implemented "Kiwi," a clinical IE package featuring both models, available at https://kiwi.clinicalnlp.org/. Conclusion: This study is among the first to develop and evaluate a comprehensive clinical IE system using open-source LLMs. Results indicate that LLaMA models outperform BiomedBERT for clinical NER and RE but with higher computational costs and lower throughputs. These findings highlight that choosing between LLMs and traditional deep learning methods for clinical IE applications should remain task-specific, taking into account both performance metrics and practical considerations such as available computing resources and the intended use case scenarios.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge