Emily Y. Chew
for the AREDS2 Deep Learning Research Group
Enhancing Large Language Models with Domain-specific Retrieval Augment Generation: A Case Study on Long-form Consumer Health Question Answering in Ophthalmology
Sep 20, 2024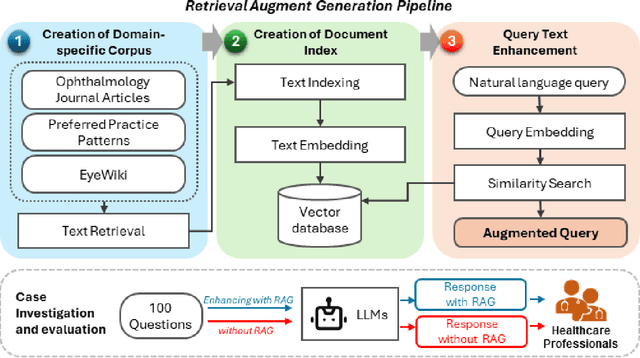

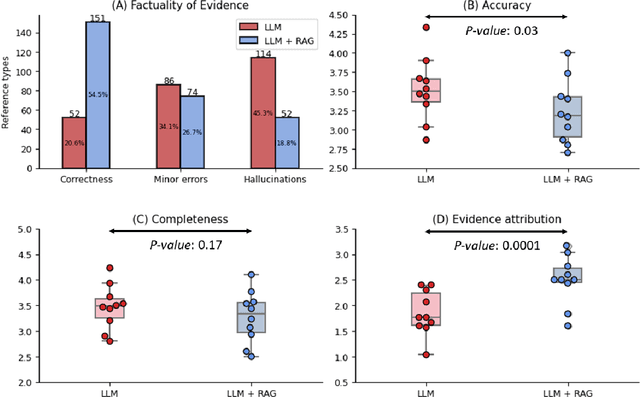

Abstract:Despite the potential of Large Language Models (LLMs) in medicine, they may generate responses lacking supporting evidence or based on hallucinated evidence. While Retrieval Augment Generation (RAG) is popular to address this issue, few studies implemented and evaluated RAG in downstream domain-specific applications. We developed a RAG pipeline with 70,000 ophthalmology-specific documents that retrieve relevant documents to augment LLMs during inference time. In a case study on long-form consumer health questions, we systematically evaluated the responses including over 500 references of LLMs with and without RAG on 100 questions with 10 healthcare professionals. The evaluation focuses on factuality of evidence, selection and ranking of evidence, attribution of evidence, and answer accuracy and completeness. LLMs without RAG provided 252 references in total. Of which, 45.3% hallucinated, 34.1% consisted of minor errors, and 20.6% were correct. In contrast, LLMs with RAG significantly improved accuracy (54.5% being correct) and reduced error rates (18.8% with minor hallucinations and 26.7% with errors). 62.5% of the top 10 documents retrieved by RAG were selected as the top references in the LLM response, with an average ranking of 4.9. The use of RAG also improved evidence attribution (increasing from 1.85 to 2.49 on a 5-point scale, P<0.001), albeit with slight decreases in accuracy (from 3.52 to 3.23, P=0.03) and completeness (from 3.47 to 3.27, P=0.17). The results demonstrate that LLMs frequently exhibited hallucinated and erroneous evidence in the responses, raising concerns for downstream applications in the medical domain. RAG substantially reduced the proportion of such evidence but encountered challenges.
Harnessing the power of longitudinal medical imaging for eye disease prognosis using Transformer-based sequence modeling
May 14, 2024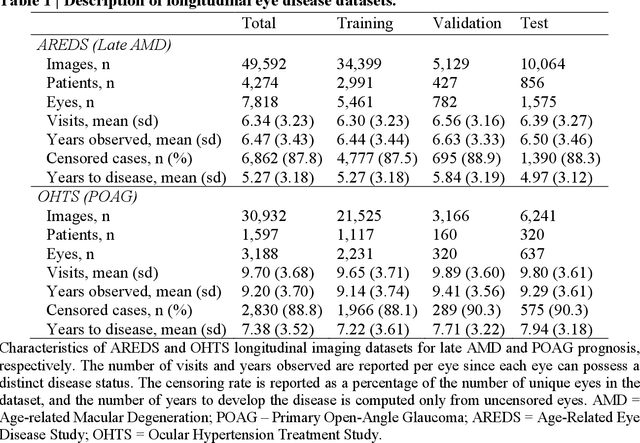
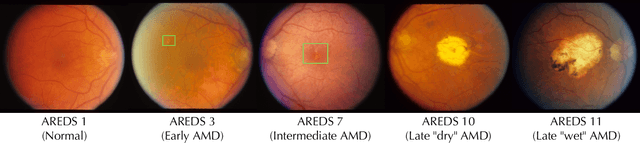
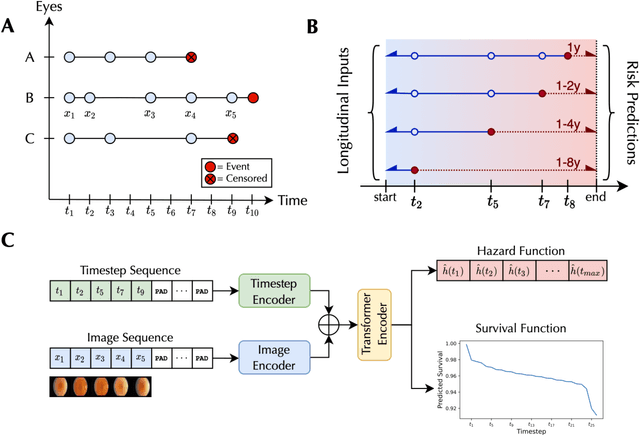
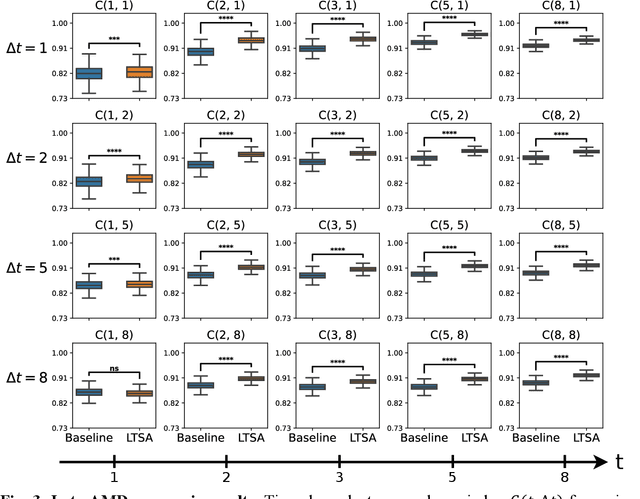
Abstract:Deep learning has enabled breakthroughs in automated diagnosis from medical imaging, with many successful applications in ophthalmology. However, standard medical image classification approaches only assess disease presence at the time of acquisition, neglecting the common clinical setting of longitudinal imaging. For slow, progressive eye diseases like age-related macular degeneration (AMD) and primary open-angle glaucoma (POAG), patients undergo repeated imaging over time to track disease progression and forecasting the future risk of developing disease is critical to properly plan treatment. Our proposed Longitudinal Transformer for Survival Analysis (LTSA) enables dynamic disease prognosis from longitudinal medical imaging, modeling the time to disease from sequences of fundus photography images captured over long, irregular time periods. Using longitudinal imaging data from the Age-Related Eye Disease Study (AREDS) and Ocular Hypertension Treatment Study (OHTS), LTSA significantly outperformed a single-image baseline in 19/20 head-to-head comparisons on late AMD prognosis and 18/20 comparisons on POAG prognosis. A temporal attention analysis also suggested that, while the most recent image is typically the most influential, prior imaging still provides additional prognostic value.
A deep learning framework for the detection and quantification of drusen and reticular pseudodrusen on optical coherence tomography
Apr 05, 2022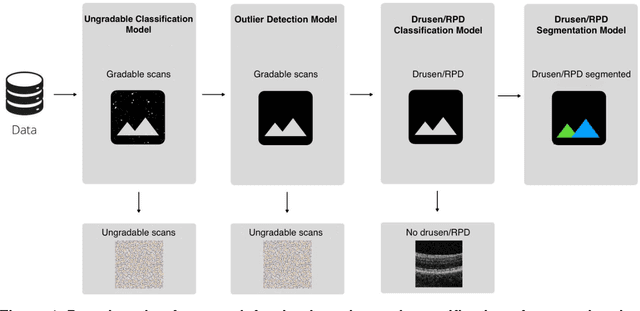
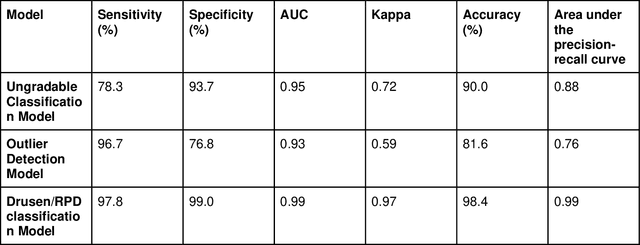
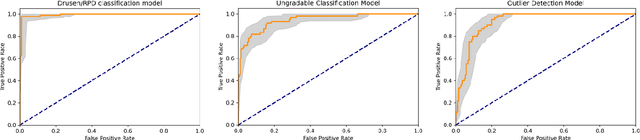
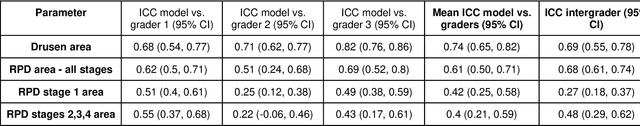
Abstract:Purpose - To develop and validate a deep learning (DL) framework for the detection and quantification of drusen and reticular pseudodrusen (RPD) on optical coherence tomography scans. Design - Development and validation of deep learning models for classification and feature segmentation. Methods - A DL framework was developed consisting of a classification model and an out-of-distribution (OOD) detection model for the identification of ungradable scans; a classification model to identify scans with drusen or RPD; and an image segmentation model to independently segment lesions as RPD or drusen. Data were obtained from 1284 participants in the UK Biobank (UKBB) with a self-reported diagnosis of age-related macular degeneration (AMD) and 250 UKBB controls. Drusen and RPD were manually delineated by five retina specialists. The main outcome measures were sensitivity, specificity, area under the ROC curve (AUC), kappa, accuracy and intraclass correlation coefficient (ICC). Results - The classification models performed strongly at their respective tasks (0.95, 0.93, and 0.99 AUC, respectively, for the ungradable scans classifier, the OOD model, and the drusen and RPD classification model). The mean ICC for drusen and RPD area vs. graders was 0.74 and 0.61, respectively, compared with 0.69 and 0.68 for intergrader agreement. FROC curves showed that the model's sensitivity was close to human performance. Conclusions - The models achieved high classification and segmentation performance, similar to human performance. Application of this robust framework will further our understanding of RPD as a separate entity from drusen in both research and clinical settings.
Multi-modal, multi-task, multi-attention deep learning detection of reticular pseudodrusen: towards automated and accessible classification of age-related macular degeneration
Nov 11, 2020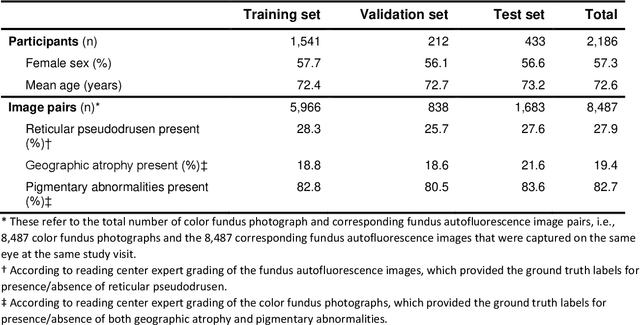
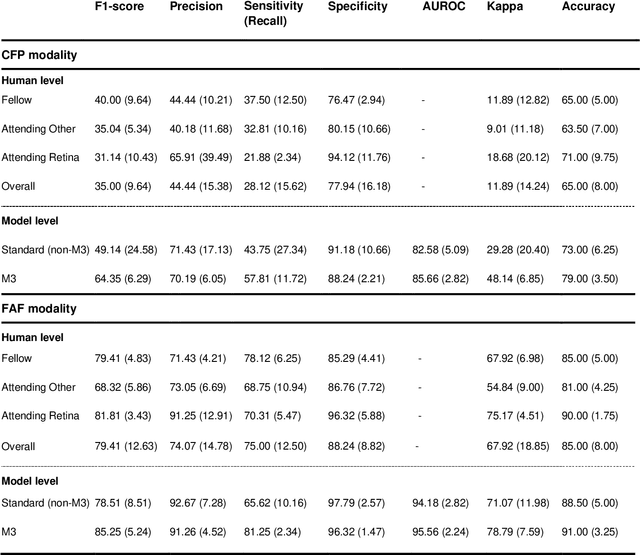
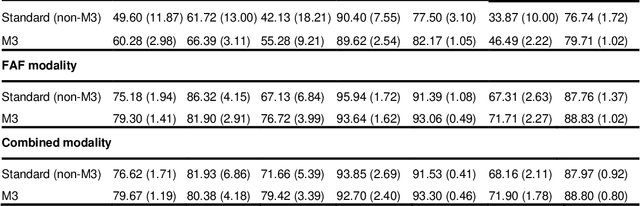
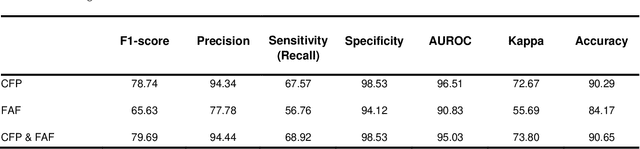
Abstract:Objective Reticular pseudodrusen (RPD), a key feature of age-related macular degeneration (AMD), are poorly detected by human experts on standard color fundus photography (CFP) and typically require advanced imaging modalities such as fundus autofluorescence (FAF). The objective was to develop and evaluate the performance of a novel 'M3' deep learning framework on RPD detection. Materials and Methods A deep learning framework M3 was developed to detect RPD presence accurately using CFP alone, FAF alone, or both, employing >8000 CFP-FAF image pairs obtained prospectively (Age-Related Eye Disease Study 2). The M3 framework includes multi-modal (detection from single or multiple image modalities), multi-task (training different tasks simultaneously to improve generalizability), and multi-attention (improving ensembled feature representation) operation. Performance on RPD detection was compared with state-of-the-art deep learning models and 13 ophthalmologists; performance on detection of two other AMD features (geographic atrophy and pigmentary abnormalities) was also evaluated. Results For RPD detection, M3 achieved area under receiver operating characteristic (AUROC) 0.832, 0.931, and 0.933 for CFP alone, FAF alone, and both, respectively. M3 performance on CFP was very substantially superior to human retinal specialists (median F1-score 0.644 versus 0.350). External validation (on Rotterdam Study, Netherlands) demonstrated high accuracy on CFP alone (AUROC 0.965). The M3 framework also accurately detected geographic atrophy and pigmentary abnormalities (AUROC 0.909 and 0.912, respectively), demonstrating its generalizability. Conclusion This study demonstrates the successful development, robust evaluation, and external validation of a novel deep learning framework that enables accessible, accurate, and automated AMD diagnosis and prognosis.
Predicting risk of late age-related macular degeneration using deep learning
Jul 19, 2020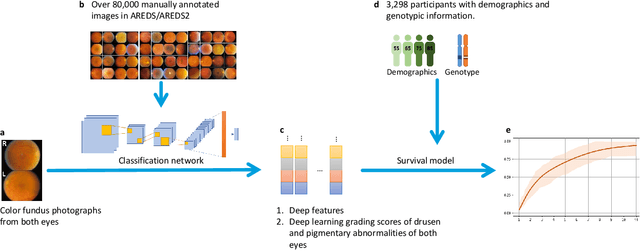
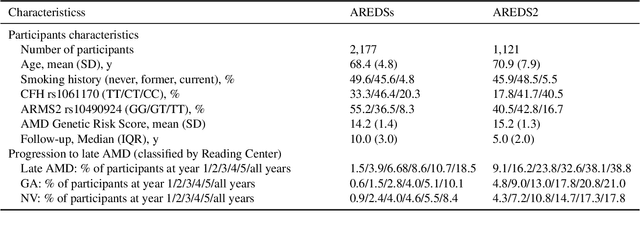
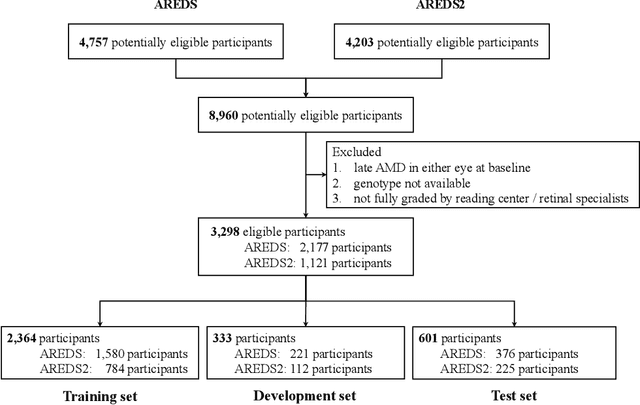
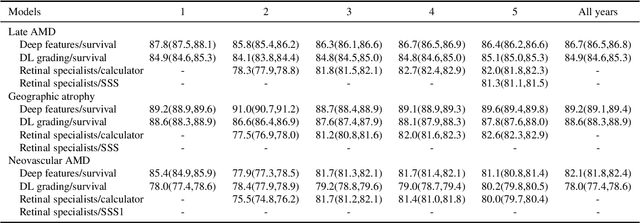
Abstract:By 2040, age-related macular degeneration (AMD) will affect approximately 288 million people worldwide. Identifying individuals at high risk of progression to late AMD, the sight-threatening stage, is critical for clinical actions, including medical interventions and timely monitoring. Although deep learning has shown promise in diagnosing/screening AMD using color fundus photographs, it remains difficult to predict individuals' risks of late AMD accurately. For both tasks, these initial deep learning attempts have remained largely unvalidated in independent cohorts. Here, we demonstrate how deep learning and survival analysis can predict the probability of progression to late AMD using 3,298 participants (over 80,000 images) from the Age-Related Eye Disease Studies AREDS and AREDS2, the largest longitudinal clinical trials in AMD. When validated against an independent test dataset of 601 participants, our model achieved high prognostic accuracy (five-year C-statistic 86.4 (95% confidence interval 86.2-86.6)) that substantially exceeded that of retinal specialists using two existing clinical standards (81.3 (81.1-81.5) and 82.0 (81.8-82.3), respectively). Interestingly, our approach offers additional strengths over the existing clinical standards in AMD prognosis (e.g., risk ascertainment above 50%) and is likely to be highly generalizable, given the breadth of training data from 82 US retinal specialty clinics. Indeed, during external validation through training on AREDS and testing on AREDS2 as an independent cohort, our model retained substantially higher prognostic accuracy than existing clinical standards. These results highlight the potential of deep learning systems to enhance clinical decision-making in AMD patients.
A deep learning approach for automated detection of geographic atrophy from color fundus photographs
Jun 07, 2019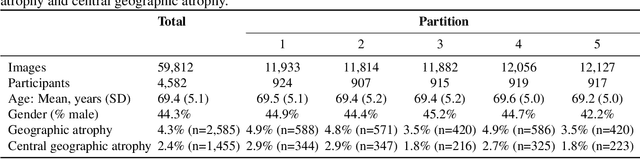
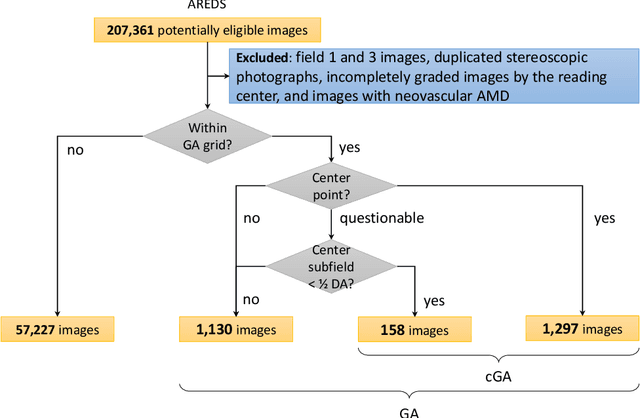
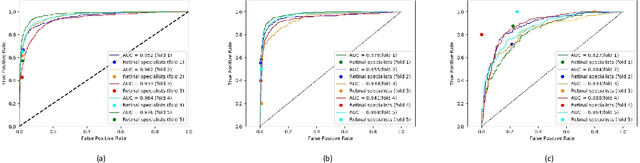
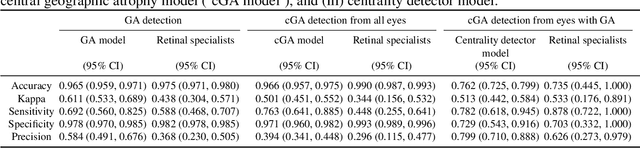
Abstract:Purpose: To assess the utility of deep learning in the detection of geographic atrophy (GA) from color fundus photographs; secondary aim to explore potential utility in detecting central GA (CGA). Design: A deep learning model was developed to detect the presence of GA in color fundus photographs, and two additional models to detect CGA in different scenarios. Participants: 59,812 color fundus photographs from longitudinal follow up of 4,582 participants in the AREDS dataset. Gold standard labels were from human expert reading center graders using a standardized protocol. Methods: A deep learning model was trained to use color fundus photographs to predict GA presence from a population of eyes with no AMD to advanced AMD. A second model was trained to predict CGA presence from the same population. A third model was trained to predict CGA presence from the subset of eyes with GA. For training and testing, 5-fold cross-validation was employed. For comparison with human clinician performance, model performance was compared with that of 88 retinal specialists. Results: The deep learning models (GA detection, CGA detection from all eyes, and centrality detection from GA eyes) had AUC of 0.933-0.976, 0.939-0.976, and 0.827-0.888, respectively. The GA detection model had accuracy, sensitivity, specificity, and precision of 0.965, 0.692, 0.978, and 0.584, respectively. The CGA detection model had equivalent values of 0.966, 0.763, 0.971, and 0.394. The centrality detection model had equivalent values of 0.762, 0.782, 0.729, and 0.799. Conclusions: A deep learning model demonstrated high accuracy for the automated detection of GA. The AUC was non-inferior to that of human retinal specialists. Deep learning approaches may also be applied to the identification of CGA. The code and pretrained models are publicly available at https://github.com/ncbi-nlp/DeepSeeNet.
A multi-task deep learning model for the classification of Age-related Macular Degeneration
Dec 02, 2018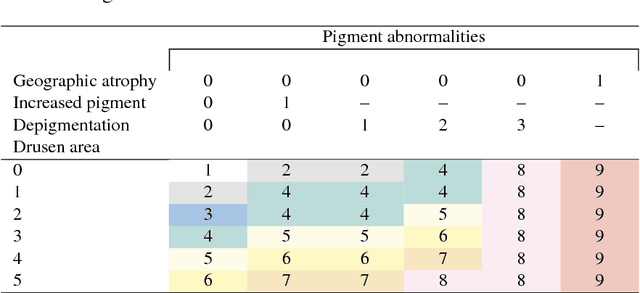
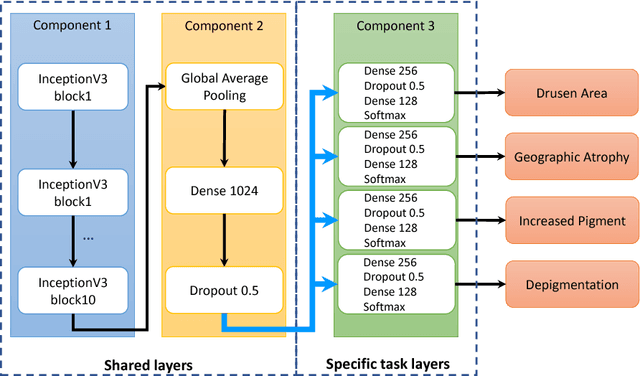
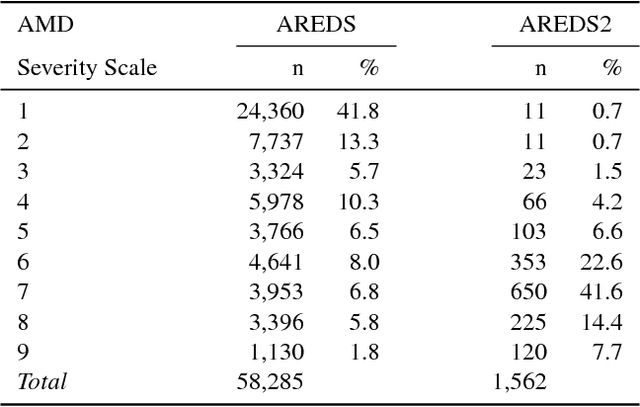
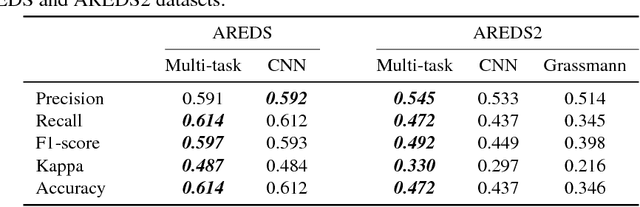
Abstract:Age-related Macular Degeneration (AMD) is a leading cause of blindness. Although the Age-Related Eye Disease Study group previously developed a 9-step AMD severity scale for manual classification of AMD severity from color fundus images, manual grading of images is time-consuming and expensive. Built on our previous work DeepSeeNet, we developed a novel deep learning model for automated classification of images into the 9-step scale. Instead of predicting the 9-step score directly, our approach simulates the reading center grading process. It first detects four AMD characteristics (drusen area, geographic atrophy, increased pigment, and depigmentation), then combines these to derive the overall 9-step score. Importantly, we applied multi-task learning techniques, which allowed us to train classification of the four characteristics in parallel, share representation, and prevent overfitting. Evaluation on two image datasets showed that the accuracy of the model exceeded the current state-of-the-art model by > 10%.
DeepSeeNet: A deep learning model for automated classification of patient-based age-related macular degeneration severity from color fundus photographs
Nov 19, 2018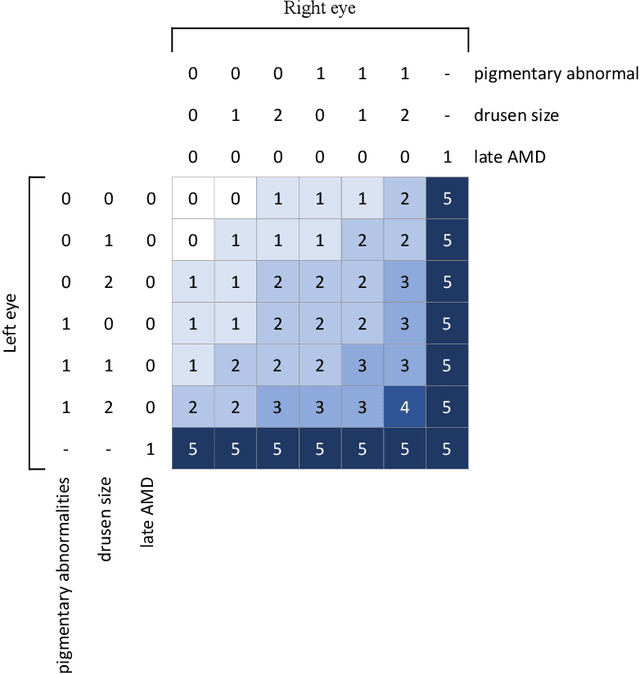
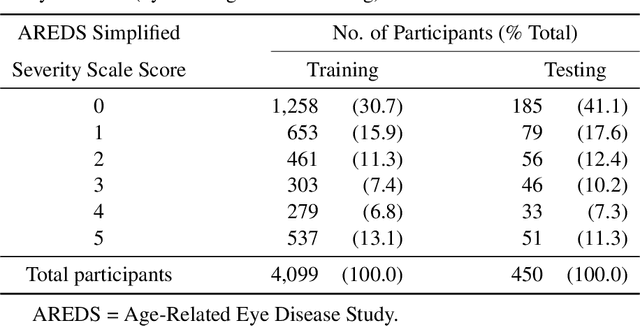
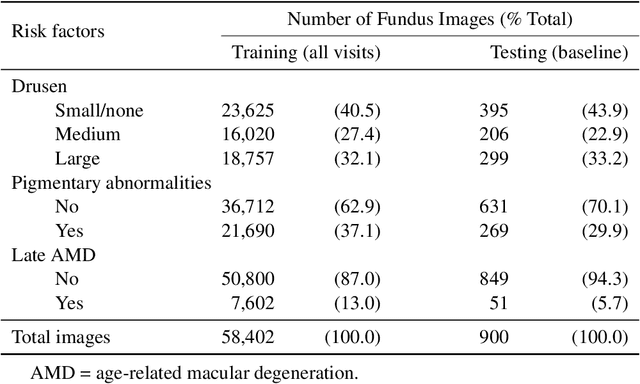
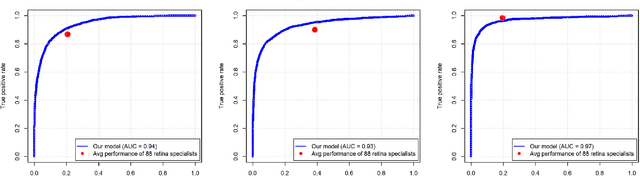
Abstract:In assessing the severity of age-related macular degeneration (AMD), the Age-Related Eye Disease Study (AREDS) Simplified Severity Scale predicts the risk of progression to late AMD. However, its manual use requires the time-consuming participation of expert practitioners. While several automated deep learning (DL) systems have been developed for classifying color fundus photographs of individual eyes by AREDS severity score, none to date has utilized a patient-based scoring system that employs images from both eyes to assign a severity score. DeepSeeNet, a DL model, was developed to classify patients automatically by the AREDS Simplified Severity Scale (score 0-5) using bilateral color fundus images. DeepSeeNet was trained on 58,402 and tested on 900 images from the longitudinal follow up of 4,549 participants from AREDS. Gold standard labels were obtained using reading center grades. DeepSeeNet simulates the human grading process by first detecting individual AMD risk factors (drusen size; pigmentary abnormalities) for each eye and then calculating a patient-based AMD severity score using the AREDS Simplified Severity Scale. DeepSeeNet performed better on patient-based, multi-class classification (accuracy=0.671; kappa=0.558) than retinal specialists (accuracy=0.599; kappa=0.467) with high AUCs in the detection of large drusen (0.94), pigmentary abnormalities (0.93) and late AMD (0.97), respectively. DeepSeeNet demonstrated high accuracy with increased transparency in the automated assignment of individual patients to AMD risk categories based on the AREDS Simplified Severity Scale. These results highlight the potential of deep learning systems to assist and enhance clinical decision-making processes in AMD patients such as early AMD detection and risk prediction for developing late AMD. DeepSeeNet is publicly available on https://github.com/ncbi-nlp/DeepSeeNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge