Aaron Y. Lee
OCTCube: A 3D foundation model for optical coherence tomography that improves cross-dataset, cross-disease, cross-device and cross-modality analysis
Aug 20, 2024Abstract:Optical coherence tomography (OCT) has become critical for diagnosing retinal diseases as it enables 3D images of the retina and optic nerve. OCT acquisition is fast, non-invasive, affordable, and scalable. Due to its broad applicability, massive numbers of OCT images have been accumulated in routine exams, making it possible to train large-scale foundation models that can generalize to various diagnostic tasks using OCT images. Nevertheless, existing foundation models for OCT only consider 2D image slices, overlooking the rich 3D structure. Here, we present OCTCube, a 3D foundation model pre-trained on 26,605 3D OCT volumes encompassing 1.62 million 2D OCT images. OCTCube is developed based on 3D masked autoencoders and exploits FlashAttention to reduce the larger GPU memory usage caused by modeling 3D volumes. OCTCube outperforms 2D models when predicting 8 retinal diseases in both inductive and cross-dataset settings, indicating that utilizing the 3D structure in the model instead of 2D data results in significant improvement. OCTCube further shows superior performance on cross-device prediction and when predicting systemic diseases, such as diabetes and hypertension, further demonstrating its strong generalizability. Finally, we propose a contrastive-self-supervised-learning-based OCT-IR pre-training framework (COIP) for cross-modality analysis on OCT and infrared retinal (IR) images, where the OCT volumes are embedded using OCTCube. We demonstrate that COIP enables accurate alignment between OCT and IR en face images. Collectively, OCTCube, a 3D OCT foundation model, demonstrates significantly better performance against 2D models on 27 out of 29 tasks and comparable performance on the other two tasks, paving the way for AI-based retinal disease diagnosis.
A deep learning framework for the detection and quantification of drusen and reticular pseudodrusen on optical coherence tomography
Apr 05, 2022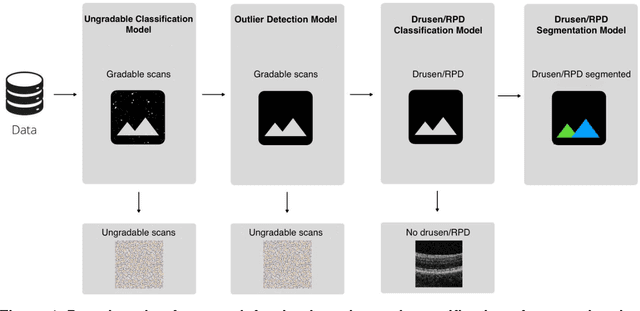
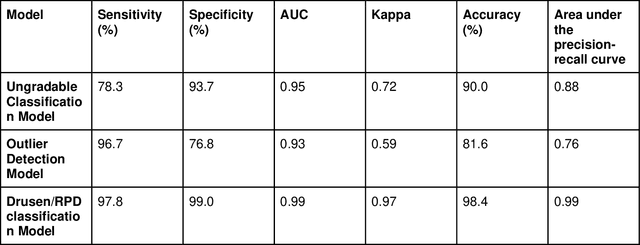
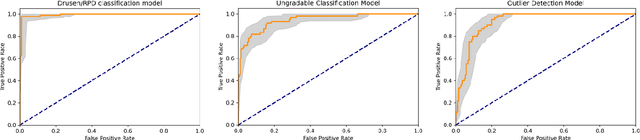
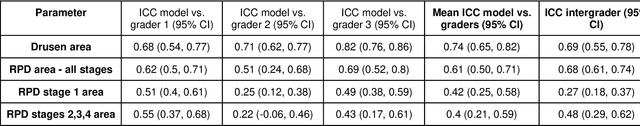
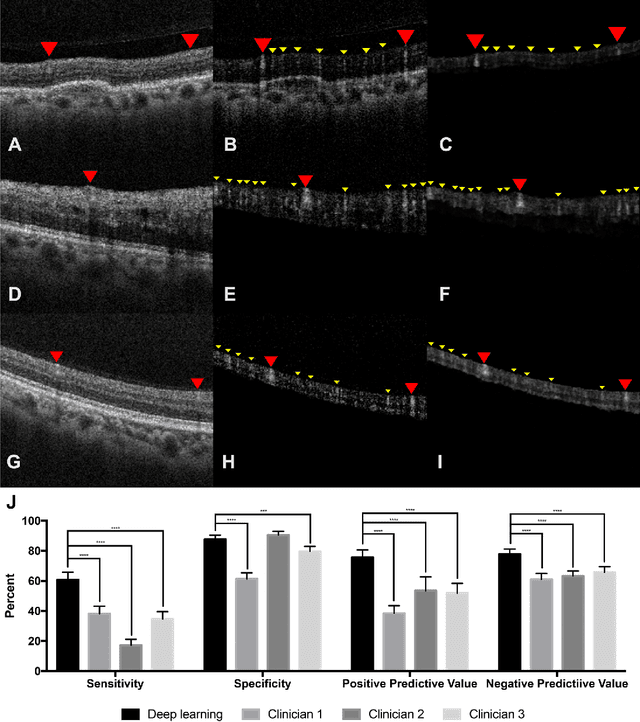
Abstract:Purpose - To develop and validate a deep learning (DL) framework for the detection and quantification of drusen and reticular pseudodrusen (RPD) on optical coherence tomography scans. Design - Development and validation of deep learning models for classification and feature segmentation. Methods - A DL framework was developed consisting of a classification model and an out-of-distribution (OOD) detection model for the identification of ungradable scans; a classification model to identify scans with drusen or RPD; and an image segmentation model to independently segment lesions as RPD or drusen. Data were obtained from 1284 participants in the UK Biobank (UKBB) with a self-reported diagnosis of age-related macular degeneration (AMD) and 250 UKBB controls. Drusen and RPD were manually delineated by five retina specialists. The main outcome measures were sensitivity, specificity, area under the ROC curve (AUC), kappa, accuracy and intraclass correlation coefficient (ICC). Results - The classification models performed strongly at their respective tasks (0.95, 0.93, and 0.99 AUC, respectively, for the ungradable scans classifier, the OOD model, and the drusen and RPD classification model). The mean ICC for drusen and RPD area vs. graders was 0.74 and 0.61, respectively, compared with 0.69 and 0.68 for intergrader agreement. FROC curves showed that the model's sensitivity was close to human performance. Conclusions - The models achieved high classification and segmentation performance, similar to human performance. Application of this robust framework will further our understanding of RPD as a separate entity from drusen in both research and clinical settings.
Forecasting Future Humphrey Visual Fields Using Deep Learning
Apr 02, 2018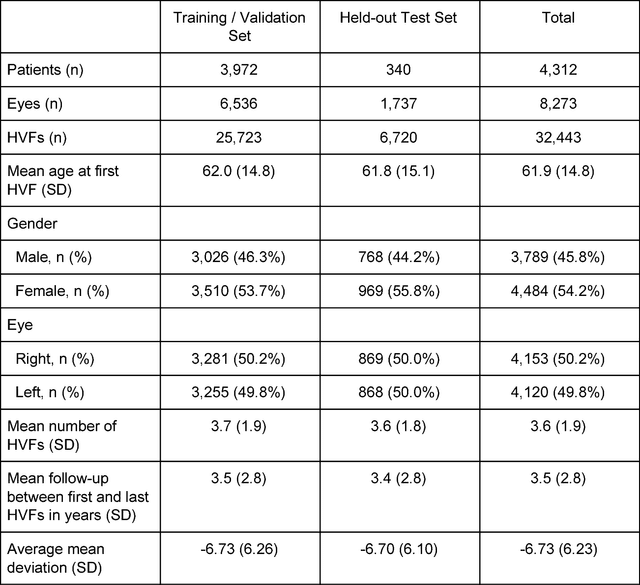
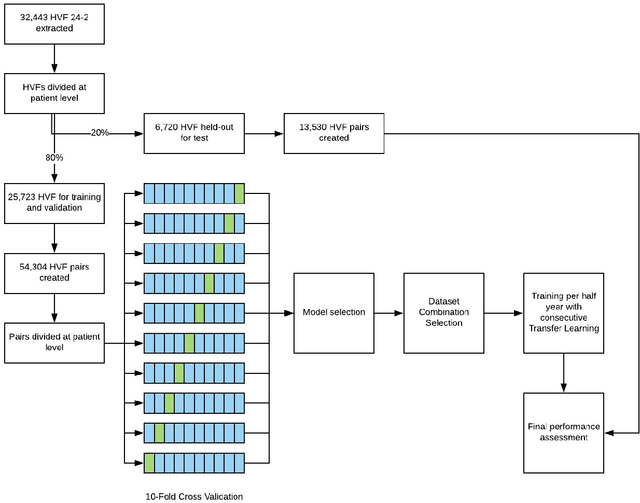

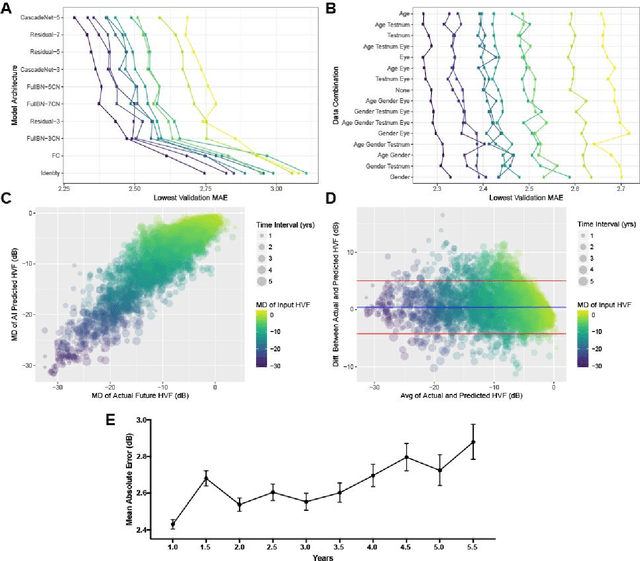
Abstract:Purpose: To determine if deep learning networks could be trained to forecast a future 24-2 Humphrey Visual Field (HVF). Participants: All patients who obtained a HVF 24-2 at the University of Washington. Methods: All datapoints from consecutive 24-2 HVFs from 1998 to 2018 were extracted from a University of Washington database. Ten-fold cross validation with a held out test set was used to develop the three main phases of model development: model architecture selection, dataset combination selection, and time-interval model training with transfer learning, to train a deep learning artificial neural network capable of generating a point-wise visual field prediction. Results: More than 1.7 million perimetry points were extracted to the hundredth decibel from 32,443 24-2 HVFs. The best performing model with 20 million trainable parameters, CascadeNet-5, was selected. The overall MAE for the test set was 2.47 dB (95% CI: 2.45 dB to 2.48 dB). The 100 fully trained models were able to successfully predict progressive field loss in glaucomatous eyes up to 5.5 years in the future with a correlation of 0.92 between the MD of predicted and actual future HVF (p < 2.2 x 10 -16 ) and an average difference of 0.41 dB. Conclusions: Using unfiltered real-world datasets, deep learning networks show an impressive ability to not only learn spatio-temporal HVF changes but also to generate predictions for future HVFs up to 5.5 years, given only a single HVF.
Generating retinal flow maps from structural optical coherence tomography with artificial intelligence
Feb 24, 2018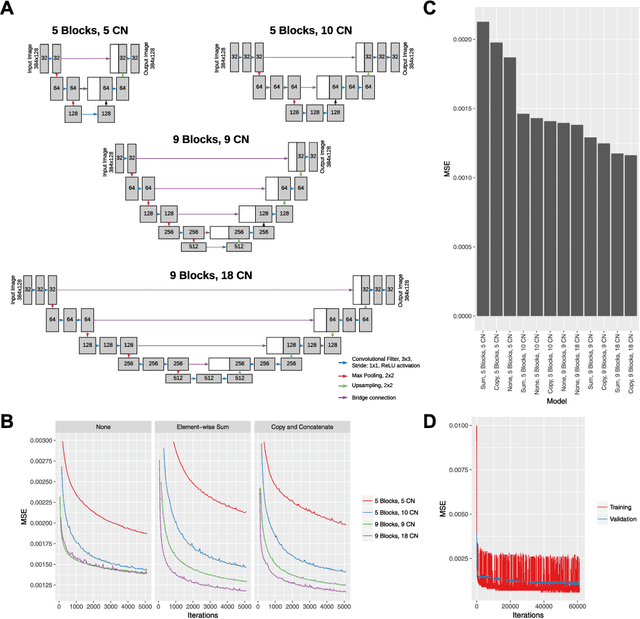

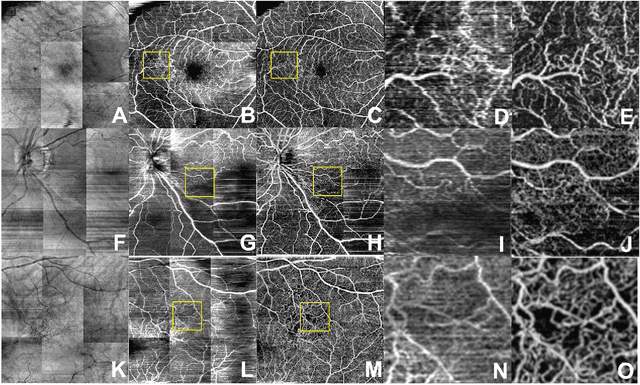
Abstract:Despite significant advances in artificial intelligence (AI) for computer vision, its application in medical imaging has been limited by the burden and limits of expert-generated labels. We used images from optical coherence tomography angiography (OCTA), a relatively new imaging modality that measures perfusion of the retinal vasculature, to train an AI algorithm to generate vasculature maps from standard structural optical coherence tomography (OCT) images of the same retinae, both exceeding the ability and bypassing the need for expert labeling. Deep learning was able to infer perfusion of microvasculature from structural OCT images with similar fidelity to OCTA and significantly better than expert clinicians (P < 0.00001). OCTA suffers from need of specialized hardware, laborious acquisition protocols, and motion artifacts; whereas our model works directly from standard OCT which are ubiquitous and quick to obtain, and allows unlocking of large volumes of previously collected standard OCT data both in existing clinical trials and clinical practice. This finding demonstrates a novel application of AI to medical imaging, whereby subtle regularities between different modalities are used to image the same body part and AI is used to generate detailed and accurate inferences of tissue function from structure imaging.
Deep learning is effective for the classification of OCT images of normal versus Age-related Macular Degeneration
Dec 15, 2016Abstract:Objective: The advent of Electronic Medical Records (EMR) with large electronic imaging databases along with advances in deep neural networks with machine learning has provided a unique opportunity to achieve milestones in automated image analysis. Optical coherence tomography (OCT) is the most commonly obtained imaging modality in ophthalmology and represents a dense and rich dataset when combined with labels derived from the EMR. We sought to determine if deep learning could be utilized to distinguish normal OCT images from images from patients with Age-related Macular Degeneration (AMD). Methods: Automated extraction of an OCT imaging database was performed and linked to clinical endpoints from the EMR. OCT macula scans were obtained by Heidelberg Spectralis, and each OCT scan was linked to EMR clinical endpoints extracted from EPIC. The central 11 images were selected from each OCT scan of two cohorts of patients: normal and AMD. Cross-validation was performed using a random subset of patients. Area under receiver operator curves (auROC) were constructed at an independent image level, macular OCT level, and patient level. Results: Of an extraction of 2.6 million OCT images linked to clinical datapoints from the EMR, 52,690 normal and 48,312 AMD macular OCT images were selected. A deep neural network was trained to categorize images as either normal or AMD. At the image level, we achieved an auROC of 92.78% with an accuracy of 87.63%. At the macula level, we achieved an auROC of 93.83% with an accuracy of 88.98%. At a patient level, we achieved an auROC of 97.45% with an accuracy of 93.45%. Peak sensitivity and specificity with optimal cutoffs were 92.64% and 93.69% respectively. Conclusions: Deep learning techniques are effective for classifying OCT images. These findings have important implications in utilizing OCT in automated screening and computer aided diagnosis tools.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge