Addie Woicik
OCTCube: A 3D foundation model for optical coherence tomography that improves cross-dataset, cross-disease, cross-device and cross-modality analysis
Aug 20, 2024Abstract:Optical coherence tomography (OCT) has become critical for diagnosing retinal diseases as it enables 3D images of the retina and optic nerve. OCT acquisition is fast, non-invasive, affordable, and scalable. Due to its broad applicability, massive numbers of OCT images have been accumulated in routine exams, making it possible to train large-scale foundation models that can generalize to various diagnostic tasks using OCT images. Nevertheless, existing foundation models for OCT only consider 2D image slices, overlooking the rich 3D structure. Here, we present OCTCube, a 3D foundation model pre-trained on 26,605 3D OCT volumes encompassing 1.62 million 2D OCT images. OCTCube is developed based on 3D masked autoencoders and exploits FlashAttention to reduce the larger GPU memory usage caused by modeling 3D volumes. OCTCube outperforms 2D models when predicting 8 retinal diseases in both inductive and cross-dataset settings, indicating that utilizing the 3D structure in the model instead of 2D data results in significant improvement. OCTCube further shows superior performance on cross-device prediction and when predicting systemic diseases, such as diabetes and hypertension, further demonstrating its strong generalizability. Finally, we propose a contrastive-self-supervised-learning-based OCT-IR pre-training framework (COIP) for cross-modality analysis on OCT and infrared retinal (IR) images, where the OCT volumes are embedded using OCTCube. We demonstrate that COIP enables accurate alignment between OCT and IR en face images. Collectively, OCTCube, a 3D OCT foundation model, demonstrates significantly better performance against 2D models on 27 out of 29 tasks and comparable performance on the other two tasks, paving the way for AI-based retinal disease diagnosis.
BLIAM: Literature-based Data Synthesis for Synergistic Drug Combination Prediction
Feb 16, 2023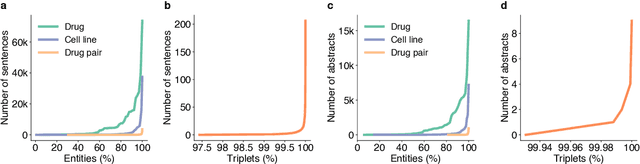
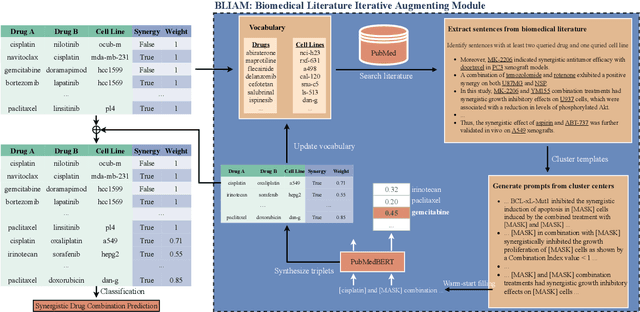
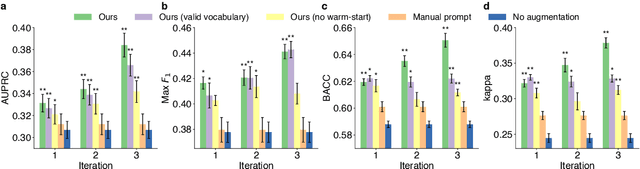
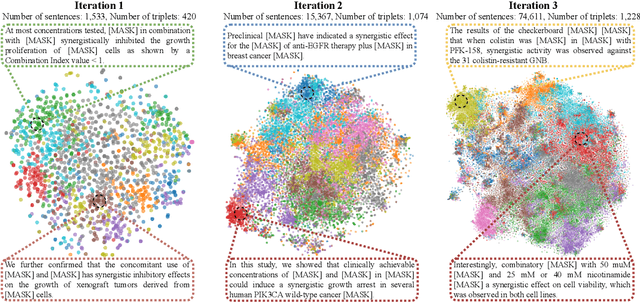
Abstract:Language models pre-trained on scientific literature corpora have substantially advanced scientific discovery by offering high-quality feature representations for downstream applications. However, these features are often not interpretable, and thus can reveal limited insights to domain experts. Instead of obtaining features from language models, we propose BLIAM, a literature-based data synthesis approach to directly generate training data points that are interpretable and model-agnostic to downstream applications. The key idea of BLIAM is to create prompts using existing training data and then use these prompts to synthesize new data points. BLIAM performs these two steps iteratively as new data points will define more informative prompts and new prompts will in turn synthesize more accurate data points. Notably, literature-based data augmentation might introduce data leakage since labels of test data points in downstream applications might have already been mentioned in the language model corpus. To prevent such leakage, we introduce GDSC-combo, a large-scale drug combination discovery dataset that was published after the biomedical language model was trained. We found that BLIAM substantially outperforms a non-augmented approach and manual prompting in this rigorous data split setting. BLIAM can be further used to synthesize data points for novel drugs and cell lines that were not even measured in biomedical experiments. In addition to the promising prediction performance, the data points synthesized by BLIAM are interpretable and model-agnostic, enabling in silico augmentation for in vitro experiments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge