Adnan Tufail
on behalf of the MACUSTAR consortium
Conditioning 3D Diffusion Models with 2D Images: Towards Standardized OCT Volumes through En Face-Informed Super-Resolution
Oct 13, 2024



Abstract:High anisotropy in volumetric medical images can lead to the inconsistent quantification of anatomical and pathological structures. Particularly in optical coherence tomography (OCT), slice spacing can substantially vary across and within datasets, studies, and clinical practices. We propose to standardize OCT volumes to less anisotropic volumes by conditioning 3D diffusion models with en face scanning laser ophthalmoscopy (SLO) imaging data, a 2D modality already commonly available in clinical practice. We trained and evaluated on data from the multicenter and multimodal MACUSTAR study. While upsampling the number of slices by a factor of 8, our method outperforms tricubic interpolation and diffusion models without en face conditioning in terms of perceptual similarity metrics. Qualitative results demonstrate improved coherence and structural similarity. Our approach allows for better informed generative decisions, potentially reducing hallucinations. We hope this work will provide the next step towards standardized high-quality volumetric imaging, enabling more consistent quantifications.
A deep learning framework for the detection and quantification of drusen and reticular pseudodrusen on optical coherence tomography
Apr 05, 2022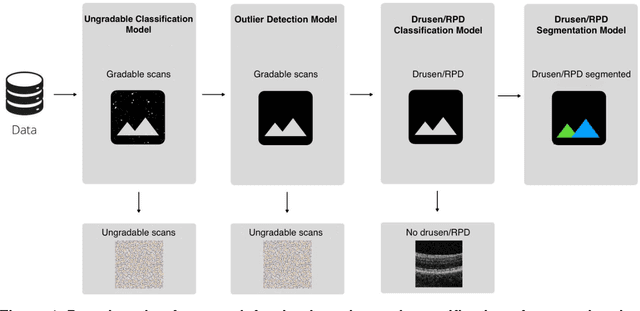
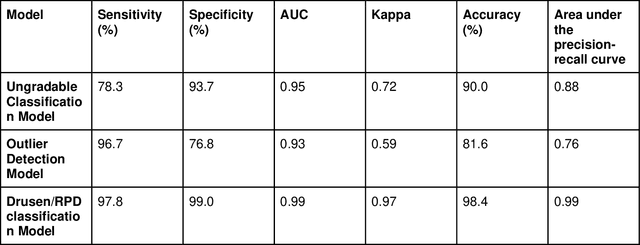
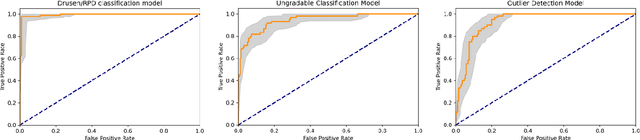
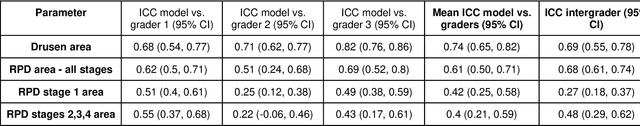
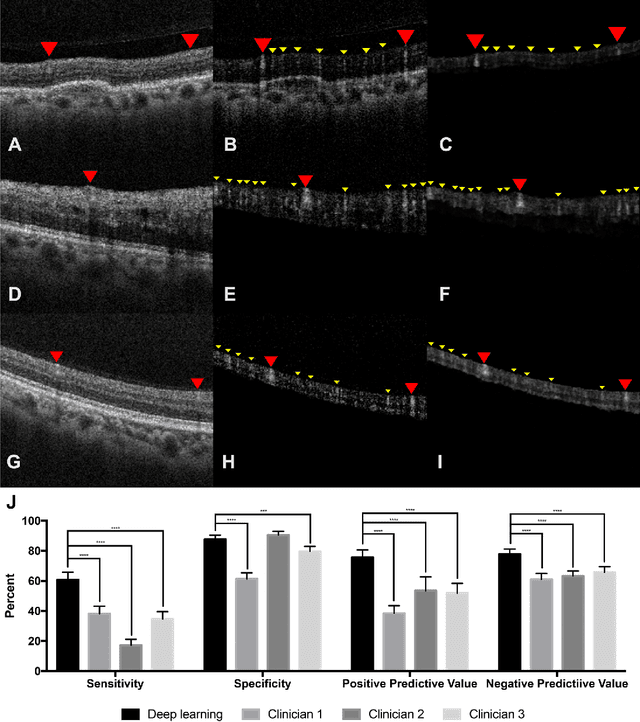
Abstract:Purpose - To develop and validate a deep learning (DL) framework for the detection and quantification of drusen and reticular pseudodrusen (RPD) on optical coherence tomography scans. Design - Development and validation of deep learning models for classification and feature segmentation. Methods - A DL framework was developed consisting of a classification model and an out-of-distribution (OOD) detection model for the identification of ungradable scans; a classification model to identify scans with drusen or RPD; and an image segmentation model to independently segment lesions as RPD or drusen. Data were obtained from 1284 participants in the UK Biobank (UKBB) with a self-reported diagnosis of age-related macular degeneration (AMD) and 250 UKBB controls. Drusen and RPD were manually delineated by five retina specialists. The main outcome measures were sensitivity, specificity, area under the ROC curve (AUC), kappa, accuracy and intraclass correlation coefficient (ICC). Results - The classification models performed strongly at their respective tasks (0.95, 0.93, and 0.99 AUC, respectively, for the ungradable scans classifier, the OOD model, and the drusen and RPD classification model). The mean ICC for drusen and RPD area vs. graders was 0.74 and 0.61, respectively, compared with 0.69 and 0.68 for intergrader agreement. FROC curves showed that the model's sensitivity was close to human performance. Conclusions - The models achieved high classification and segmentation performance, similar to human performance. Application of this robust framework will further our understanding of RPD as a separate entity from drusen in both research and clinical settings.
Unsupervised cross domain learning with applications to 7 layer segmentation of OCTs
Nov 23, 2021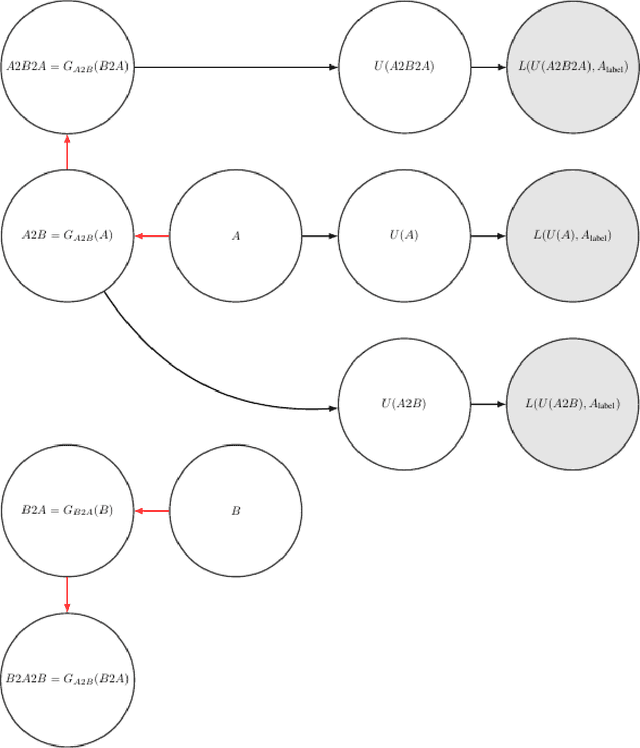
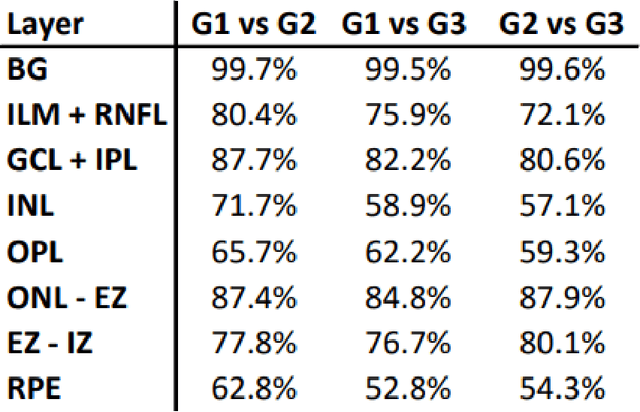
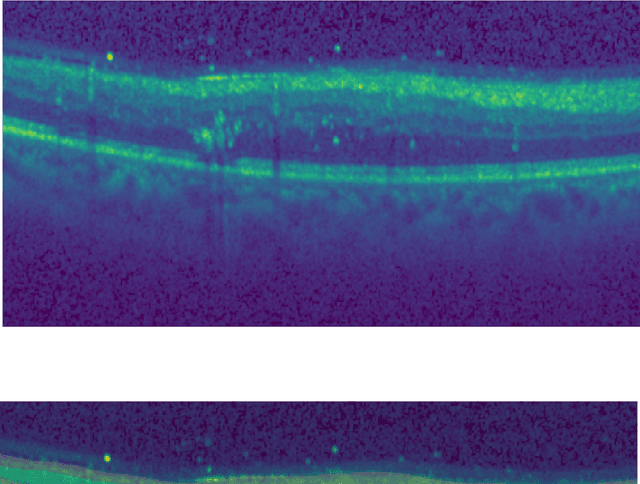
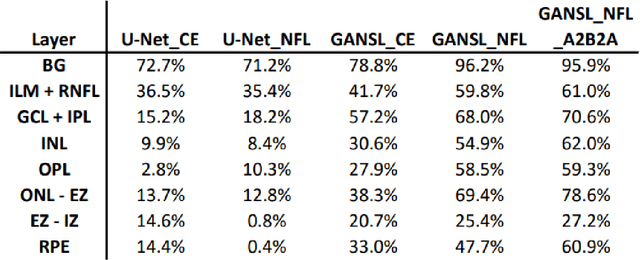
Abstract:Unsupervised cross domain adaptation for OCT 7 layer segmentation and other medical applications where labeled training data is only available in a source domain and unavailable in the target domain. Our proposed method helps generalize of deep learning to many areas in the medical field where labeled training data are expensive and time consuming to acquire or where target domains are too novel to have had labelling.
Generating retinal flow maps from structural optical coherence tomography with artificial intelligence
Feb 24, 2018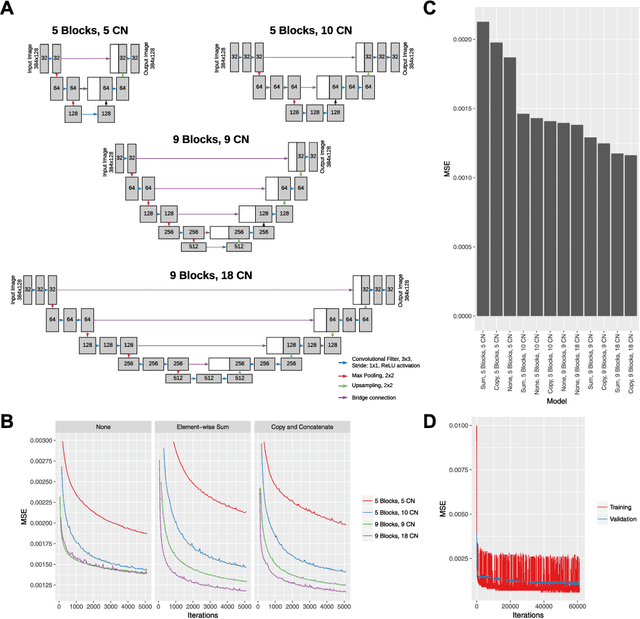

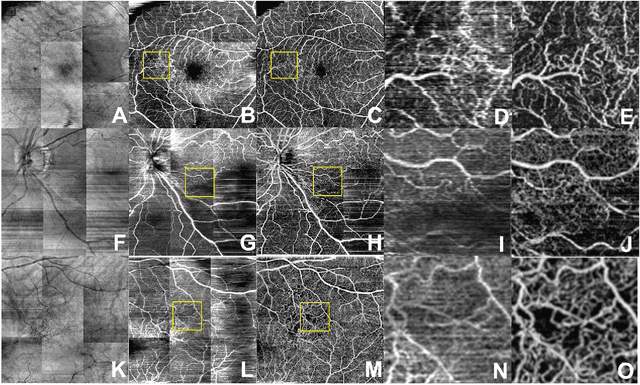
Abstract:Despite significant advances in artificial intelligence (AI) for computer vision, its application in medical imaging has been limited by the burden and limits of expert-generated labels. We used images from optical coherence tomography angiography (OCTA), a relatively new imaging modality that measures perfusion of the retinal vasculature, to train an AI algorithm to generate vasculature maps from standard structural optical coherence tomography (OCT) images of the same retinae, both exceeding the ability and bypassing the need for expert labeling. Deep learning was able to infer perfusion of microvasculature from structural OCT images with similar fidelity to OCTA and significantly better than expert clinicians (P < 0.00001). OCTA suffers from need of specialized hardware, laborious acquisition protocols, and motion artifacts; whereas our model works directly from standard OCT which are ubiquitous and quick to obtain, and allows unlocking of large volumes of previously collected standard OCT data both in existing clinical trials and clinical practice. This finding demonstrates a novel application of AI to medical imaging, whereby subtle regularities between different modalities are used to image the same body part and AI is used to generate detailed and accurate inferences of tissue function from structure imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge