Aaron Lee
Unsupervised cross domain learning with applications to 7 layer segmentation of OCTs
Nov 23, 2021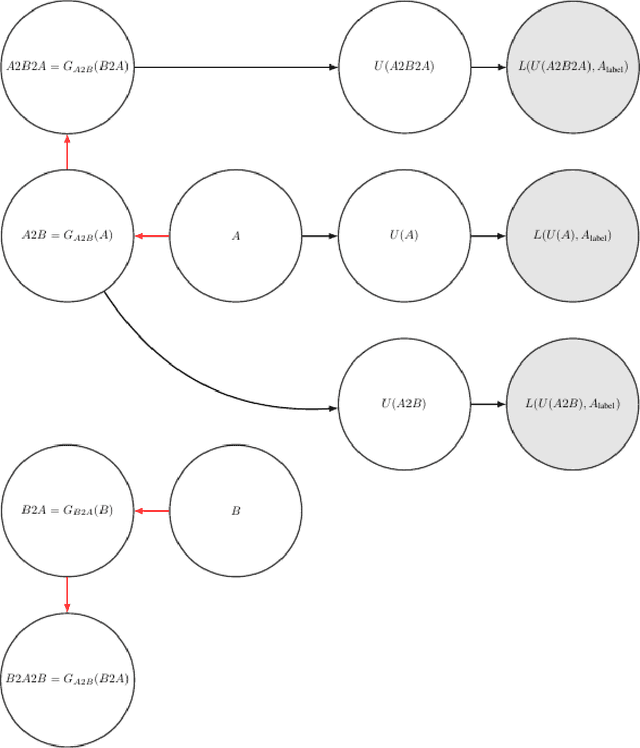
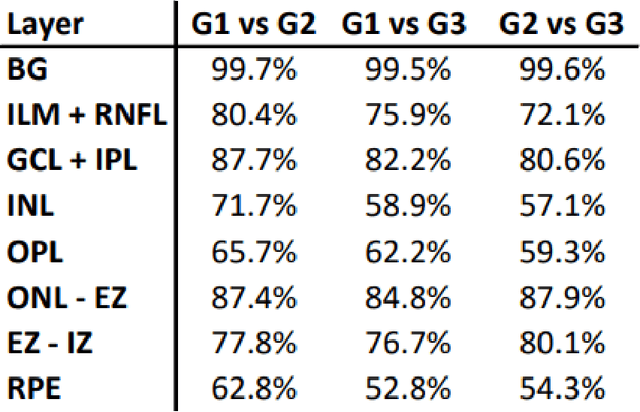
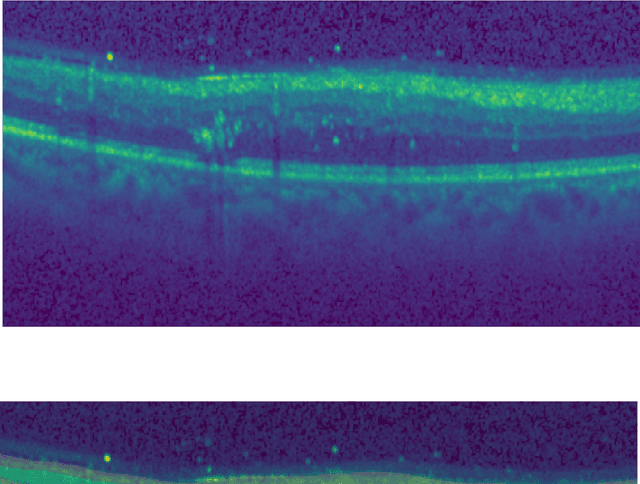
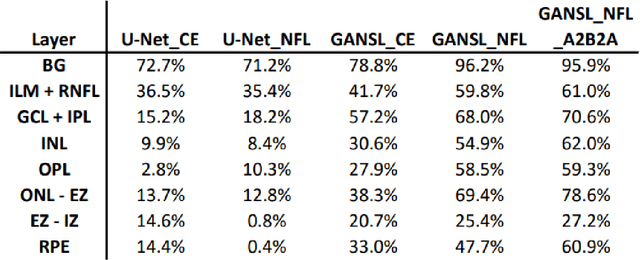
Abstract:Unsupervised cross domain adaptation for OCT 7 layer segmentation and other medical applications where labeled training data is only available in a source domain and unavailable in the target domain. Our proposed method helps generalize of deep learning to many areas in the medical field where labeled training data are expensive and time consuming to acquire or where target domains are too novel to have had labelling.
Real-time Prediction of Segmentation Quality
Jun 16, 2018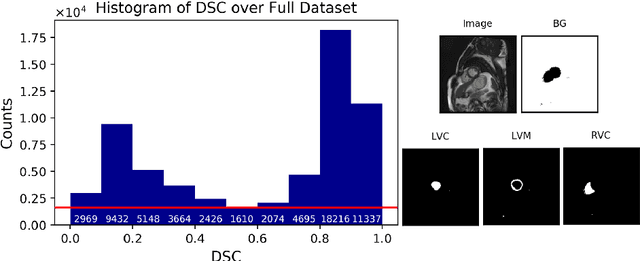
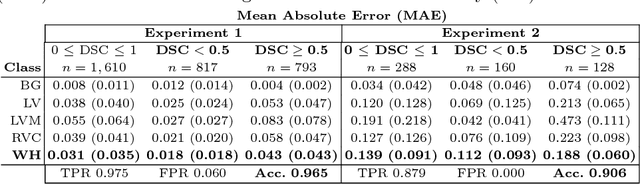
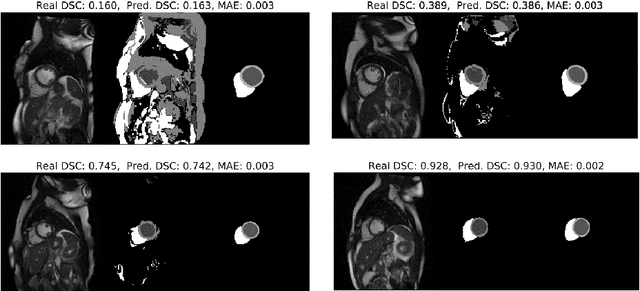
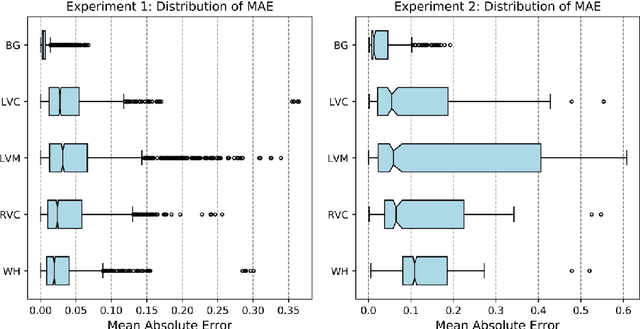
Abstract:Recent advances in deep learning based image segmentation methods have enabled real-time performance with human-level accuracy. However, occasionally even the best method fails due to low image quality, artifacts or unexpected behaviour of black box algorithms. Being able to predict segmentation quality in the absence of ground truth is of paramount importance in clinical practice, but also in large-scale studies to avoid the inclusion of invalid data in subsequent analysis. In this work, we propose two approaches of real-time automated quality control for cardiovascular MR segmentations using deep learning. First, we train a neural network on 12,880 samples to predict Dice Similarity Coefficients (DSC) on a per-case basis. We report a mean average error (MAE) of 0.03 on 1,610 test samples and 97% binary classification accuracy for separating low and high quality segmentations. Secondly, in the scenario where no manually annotated data is available, we train a network to predict DSC scores from estimated quality obtained via a reverse testing strategy. We report an MAE=0.14 and 91% binary classification accuracy for this case. Predictions are obtained in real-time which, when combined with real-time segmentation methods, enables instant feedback on whether an acquired scan is analysable while the patient is still in the scanner. This further enables new applications of optimising image acquisition towards best possible analysis results.
ACO Implementation for Sequence Alignment with Genetic Algorithms
Jun 04, 2014



Abstract:In this paper, we implement Ant Colony Optimization (ACO) for sequence alignment. ACO is a meta-heuristic recently developed for nearest neighbor approximations in large, NP-hard search spaces. Here we use a genetic algorithm approach to evolve the best parameters for an ACO designed to align two sequences. We then used the best parameters found to interpolate approximate optimal parameters for a given string length within a range. The basis of our comparison is the alignment given by the Needleman-Wunsch algorithm. We found that ACO can indeed be applied to sequence alignment. While it is computationally expensive compared to other equivalent algorithms, it is a promising algorithm that can be readily applied to a variety of other biological problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge