Nay Aung
William Harvey Research Institute, Queen Mary University of London, Barts Heart Centre, London
SAGCNet: Spatial-Aware Graph Completion Network for Missing Slice Imputation in Population CMR Imaging
Aug 09, 2025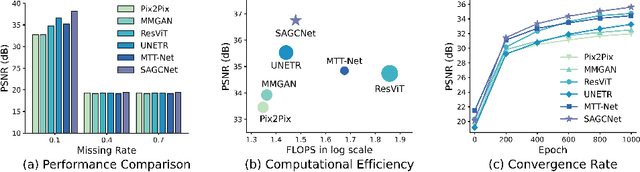
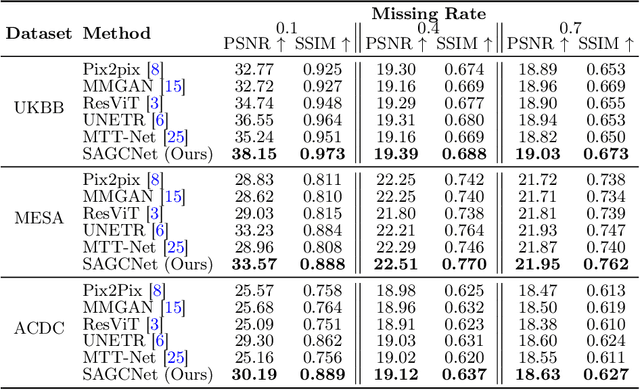
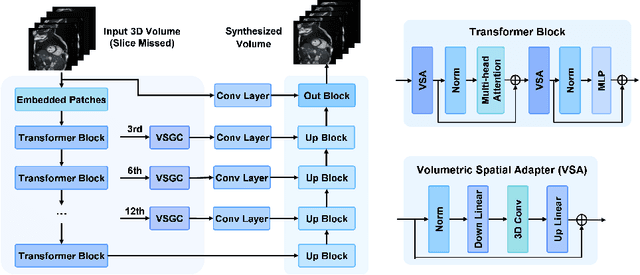
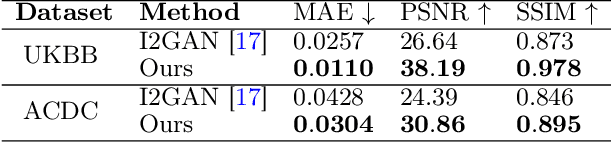
Abstract:Magnetic resonance imaging (MRI) provides detailed soft-tissue characteristics that assist in disease diagnosis and screening. However, the accuracy of clinical practice is often hindered by missing or unusable slices due to various factors. Volumetric MRI synthesis methods have been developed to address this issue by imputing missing slices from available ones. The inherent 3D nature of volumetric MRI data, such as cardiac magnetic resonance (CMR), poses significant challenges for missing slice imputation approaches, including (1) the difficulty of modeling local inter-slice correlations and dependencies of volumetric slices, and (2) the limited exploration of crucial 3D spatial information and global context. In this study, to mitigate these issues, we present Spatial-Aware Graph Completion Network (SAGCNet) to overcome the dependency on complete volumetric data, featuring two main innovations: (1) a volumetric slice graph completion module that incorporates the inter-slice relationships into a graph structure, and (2) a volumetric spatial adapter component that enables our model to effectively capture and utilize various forms of 3D spatial context. Extensive experiments on cardiac MRI datasets demonstrate that SAGCNet is capable of synthesizing absent CMR slices, outperforming competitive state-of-the-art MRI synthesis methods both quantitatively and qualitatively. Notably, our model maintains superior performance even with limited slice data.
Debiasing Cardiac Imaging with Controlled Latent Diffusion Models
Mar 28, 2024

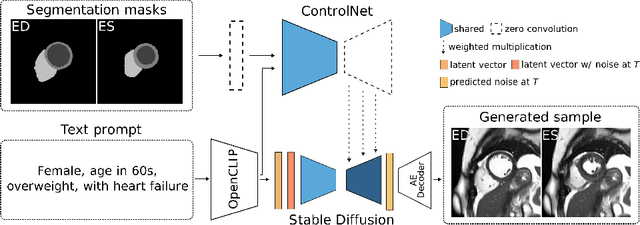

Abstract:The progress in deep learning solutions for disease diagnosis and prognosis based on cardiac magnetic resonance imaging is hindered by highly imbalanced and biased training data. To address this issue, we propose a method to alleviate imbalances inherent in datasets through the generation of synthetic data based on sensitive attributes such as sex, age, body mass index, and health condition. We adopt ControlNet based on a denoising diffusion probabilistic model to condition on text assembled from patient metadata and cardiac geometry derived from segmentation masks using a large-cohort study, specifically, the UK Biobank. We assess our method by evaluating the realism of the generated images using established quantitative metrics. Furthermore, we conduct a downstream classification task aimed at debiasing a classifier by rectifying imbalances within underrepresented groups through synthetically generated samples. Our experiments demonstrate the effectiveness of the proposed approach in mitigating dataset imbalances, such as the scarcity of younger patients or individuals with normal BMI level suffering from heart failure. This work represents a major step towards the adoption of synthetic data for the development of fair and generalizable models for medical classification tasks. Notably, we conduct all our experiments using a single, consumer-level GPU to highlight the feasibility of our approach within resource-constrained environments. Our code is available at https://github.com/faildeny/debiasing-cardiac-mri.
Fully Automated Myocardial Strain Estimation from CMR Tagged Images using a Deep Learning Framework in the UK Biobank
Apr 15, 2020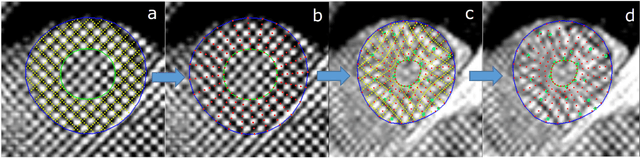
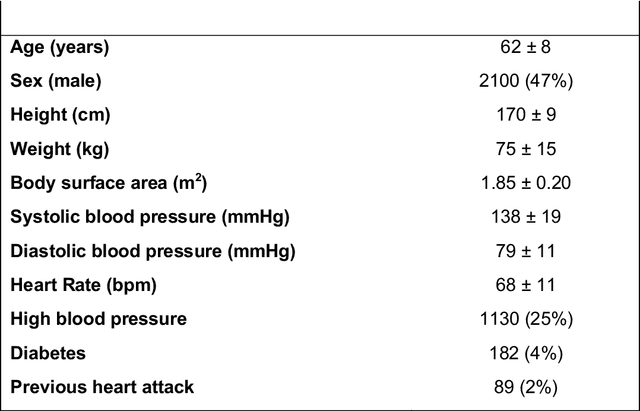
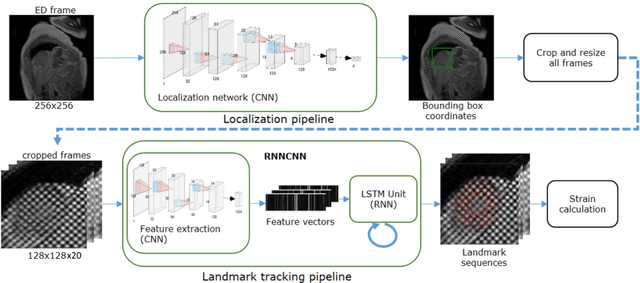
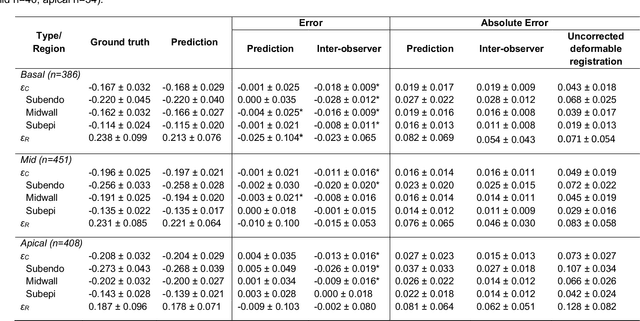
Abstract:Purpose: To demonstrate the feasibility and performance of a fully automated deep learning framework to estimate myocardial strain from short-axis cardiac magnetic resonance tagged images. Methods and Materials: In this retrospective cross-sectional study, 4508 cases from the UK Biobank were split randomly into 3244 training and 812 validation cases, and 452 test cases. Ground truth myocardial landmarks were defined and tracked by manual initialization and correction of deformable image registration using previously validated software with five readers. The fully automatic framework consisted of 1) a convolutional neural network (CNN) for localization, and 2) a combination of a recurrent neural network (RNN) and a CNN to detect and track the myocardial landmarks through the image sequence for each slice. Radial and circumferential strain were then calculated from the motion of the landmarks and averaged on a slice basis. Results: Within the test set, myocardial end-systolic circumferential Green strain errors were -0.001 +/- 0.025, -0.001 +/- 0.021, and 0.004 +/- 0.035 in basal, mid, and apical slices respectively (mean +/- std. dev. of differences between predicted and manual strain). The framework reproduced significant reductions in circumferential strain in diabetics, hypertensives, and participants with previous heart attack. Typical processing time was ~260 frames (~13 slices) per second on an NVIDIA Tesla K40 with 12GB RAM, compared with 6-8 minutes per slice for the manual analysis. Conclusions: The fully automated RNNCNN framework for analysis of myocardial strain enabled unbiased strain evaluation in a high-throughput workflow, with similar ability to distinguish impairment due to diabetes, hypertension, and previous heart attack.
* accepted in Radiology Cardiothoracic Imaging
Improving the generalizability of convolutional neural network-based segmentation on CMR images
Jul 03, 2019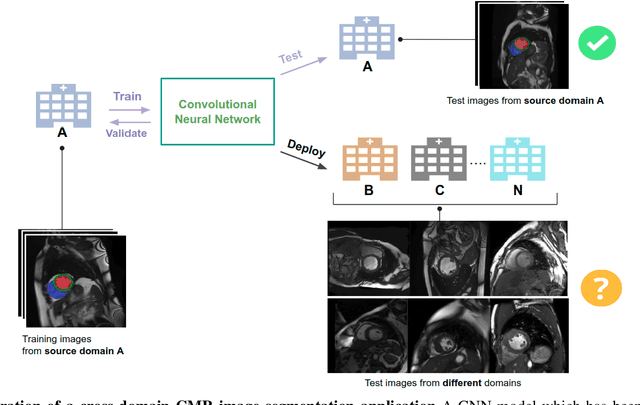
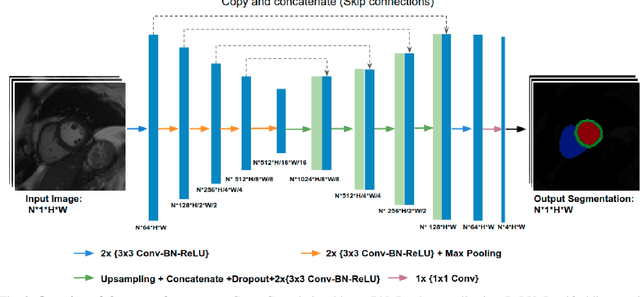
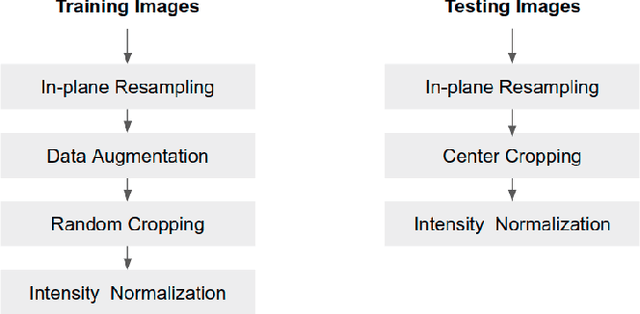
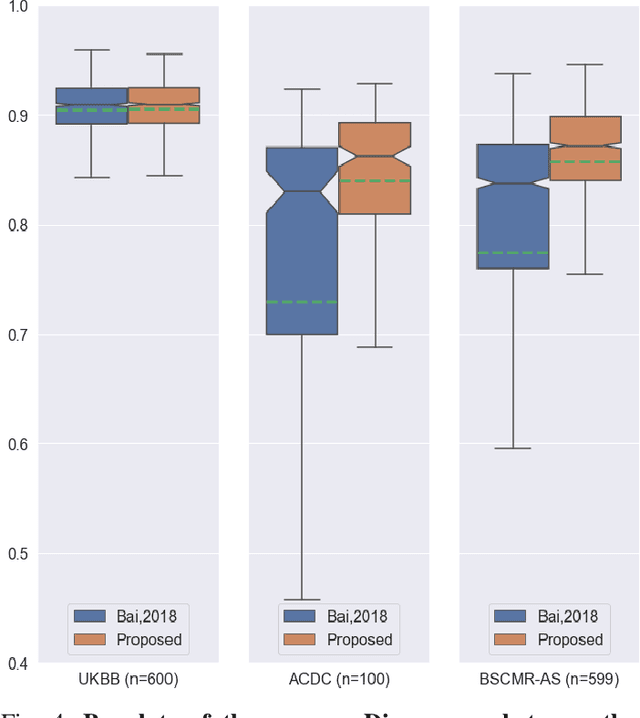
Abstract:Convolutional neural network (CNN) based segmentation methods provide an efficient and automated way for clinicians to assess the structure and function of the heart in cardiac MR images. While CNNs can generally perform the segmentation tasks with high accuracy when training and test images come from the same domain (e.g. same scanner or site), their performance often degrades dramatically on images from different scanners or clinical sites. We propose a simple yet effective way for improving the network generalization ability by carefully designing data normalization and augmentation strategies to accommodate common scenarios in multi-site, multi-scanner clinical imaging data sets. We demonstrate that a neural network trained on a single-site single-scanner dataset from the UK Biobank can be successfully applied to segmenting cardiac MR images across different sites and different scanners without substantial loss of accuracy. Specifically, the method was trained on a large set of 3,975 subjects from the UK Biobank. It was then directly tested on 600 different subjects from the UK Biobank for intra-domain testing and two other sets for cross-domain testing: the ACDC dataset (100 subjects, 1 site, 2 scanners) and the BSCMR-AS dataset (599 subjects, 6 sites, 9 scanners). The proposed method produces promising segmentation results on the UK Biobank test set which are comparable to previously reported values in the literature, while also performing well on cross-domain test sets, achieving a mean Dice metric of 0.90 for the left ventricle, 0.81 for the myocardium and 0.82 for the right ventricle on the ACDC dataset; and 0.89 for the left ventricle, 0.83 for the myocardium on the BSCMR-AS dataset. The proposed method offers a potential solution to improve CNN-based model generalizability for the cross-scanner and cross-site cardiac MR image segmentation task.
Automated Quality Control in Image Segmentation: Application to the UK Biobank Cardiac MR Imaging Study
Jan 27, 2019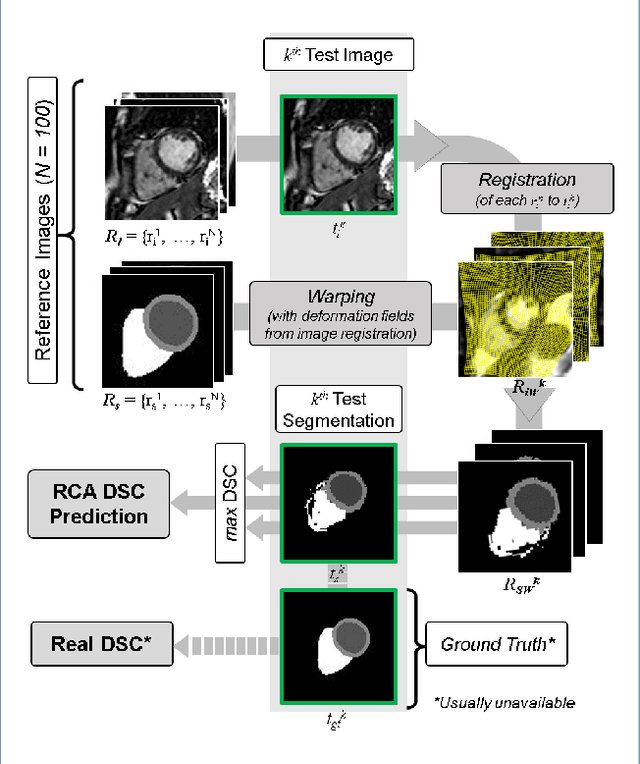
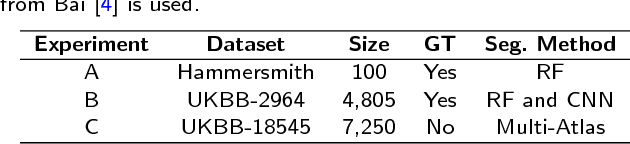
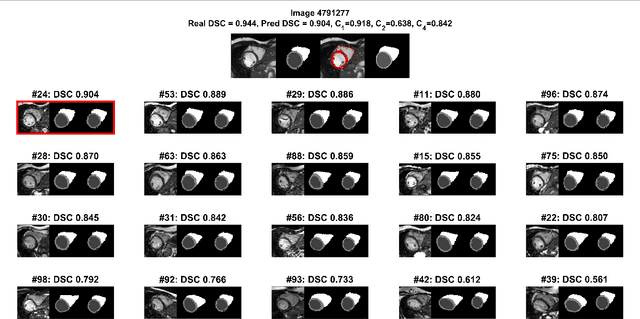
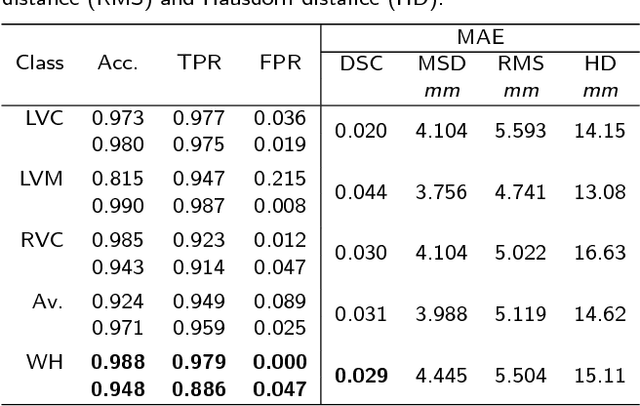
Abstract:Background: The trend towards large-scale studies including population imaging poses new challenges in terms of quality control (QC). This is a particular issue when automatic processing tools, e.g. image segmentation methods, are employed to derive quantitative measures or biomarkers for later analyses. Manual inspection and visual QC of each segmentation isn't feasible at large scale. However, it's important to be able to automatically detect when a segmentation method fails so as to avoid inclusion of wrong measurements into subsequent analyses which could lead to incorrect conclusions. Methods: To overcome this challenge, we explore an approach for predicting segmentation quality based on Reverse Classification Accuracy, which enables us to discriminate between successful and failed segmentations on a per-cases basis. We validate this approach on a new, large-scale manually-annotated set of 4,800 cardiac magnetic resonance scans. We then apply our method to a large cohort of 7,250 cardiac MRI on which we have performed manual QC. Results: We report results used for predicting segmentation quality metrics including Dice Similarity Coefficient (DSC) and surface-distance measures. As initial validation, we present data for 400 scans demonstrating 99% accuracy for classifying low and high quality segmentations using predicted DSC scores. As further validation we show high correlation between real and predicted scores and 95% classification accuracy on 4,800 scans for which manual segmentations were available. We mimic real-world application of the method on 7,250 cardiac MRI where we show good agreement between predicted quality metrics and manual visual QC scores. Conclusions: We show that RCA has the potential for accurate and fully automatic segmentation QC on a per-case basis in the context of large-scale population imaging as in the UK Biobank Imaging Study.
Real-time Prediction of Segmentation Quality
Jun 16, 2018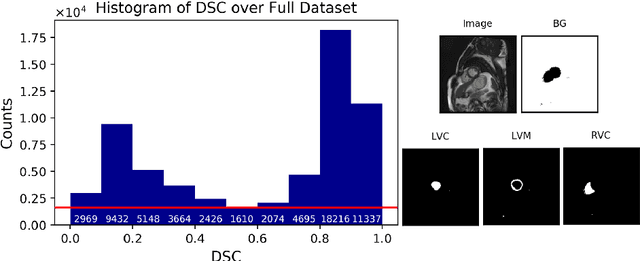
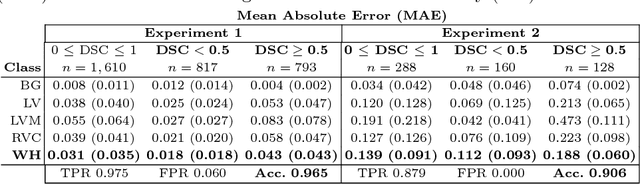
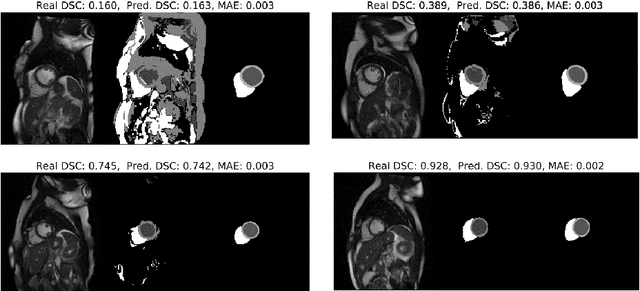
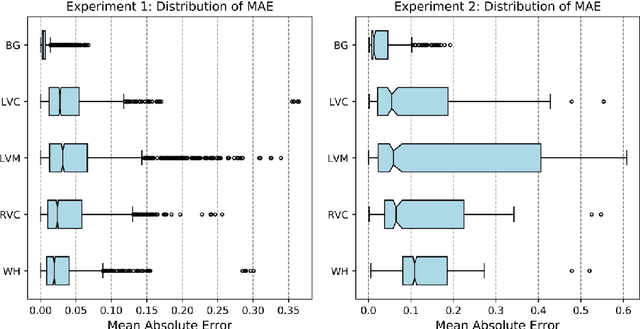
Abstract:Recent advances in deep learning based image segmentation methods have enabled real-time performance with human-level accuracy. However, occasionally even the best method fails due to low image quality, artifacts or unexpected behaviour of black box algorithms. Being able to predict segmentation quality in the absence of ground truth is of paramount importance in clinical practice, but also in large-scale studies to avoid the inclusion of invalid data in subsequent analysis. In this work, we propose two approaches of real-time automated quality control for cardiovascular MR segmentations using deep learning. First, we train a neural network on 12,880 samples to predict Dice Similarity Coefficients (DSC) on a per-case basis. We report a mean average error (MAE) of 0.03 on 1,610 test samples and 97% binary classification accuracy for separating low and high quality segmentations. Secondly, in the scenario where no manually annotated data is available, we train a network to predict DSC scores from estimated quality obtained via a reverse testing strategy. We report an MAE=0.14 and 91% binary classification accuracy for this case. Predictions are obtained in real-time which, when combined with real-time segmentation methods, enables instant feedback on whether an acquired scan is analysable while the patient is still in the scanner. This further enables new applications of optimising image acquisition towards best possible analysis results.
Automated cardiovascular magnetic resonance image analysis with fully convolutional networks
May 22, 2018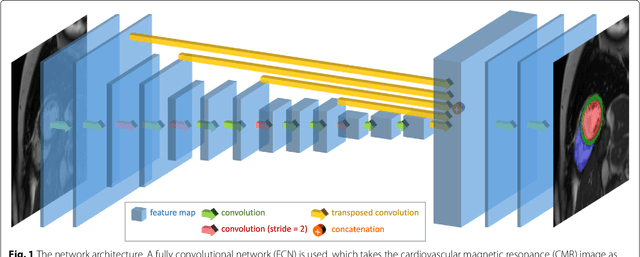

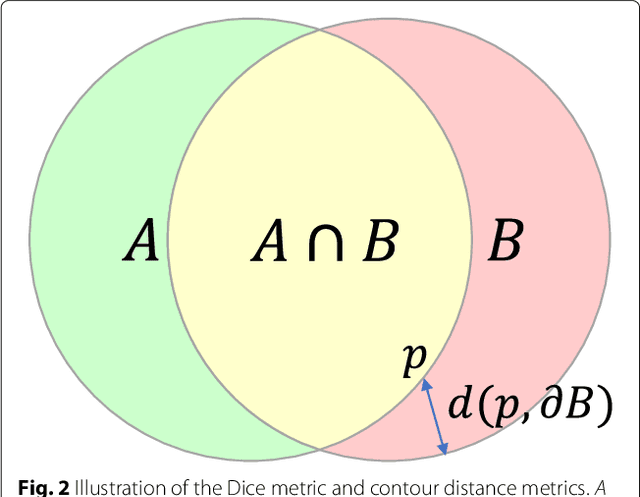
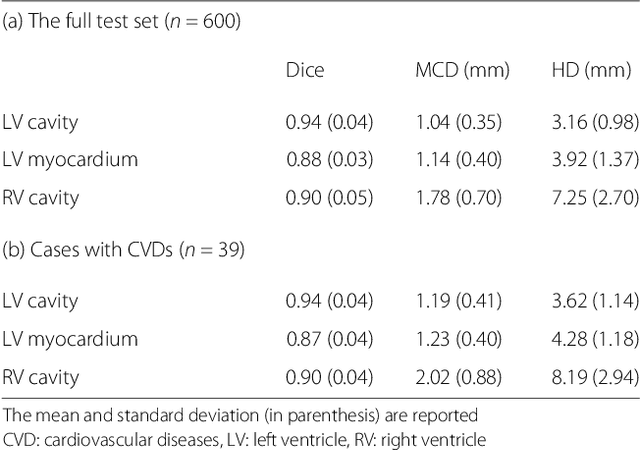
Abstract:Cardiovascular magnetic resonance (CMR) imaging is a standard imaging modality for assessing cardiovascular diseases (CVDs), the leading cause of death globally. CMR enables accurate quantification of the cardiac chamber volume, ejection fraction and myocardial mass, providing information for diagnosis and monitoring of CVDs. However, for years, clinicians have been relying on manual approaches for CMR image analysis, which is time consuming and prone to subjective errors. It is a major clinical challenge to automatically derive quantitative and clinically relevant information from CMR images. Deep neural networks have shown a great potential in image pattern recognition and segmentation for a variety of tasks. Here we demonstrate an automated analysis method for CMR images, which is based on a fully convolutional network (FCN). The network is trained and evaluated on a large-scale dataset from the UK Biobank, consisting of 4,875 subjects with 93,500 pixelwise annotated images. The performance of the method has been evaluated using a number of technical metrics, including the Dice metric, mean contour distance and Hausdorff distance, as well as clinically relevant measures, including left ventricle (LV) end-diastolic volume (LVEDV) and end-systolic volume (LVESV), LV mass (LVM); right ventricle (RV) end-diastolic volume (RVEDV) and end-systolic volume (RVESV). By combining FCN with a large-scale annotated dataset, the proposed automated method achieves a high performance on par with human experts in segmenting the LV and RV on short-axis CMR images and the left atrium (LA) and right atrium (RA) on long-axis CMR images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge