Steffen E Petersen
Neural Implicit Heart Coordinates: 3D cardiac shape reconstruction from sparse segmentations
Dec 23, 2025Abstract:Accurate reconstruction of cardiac anatomy from sparse clinical images remains a major challenge in patient-specific modeling. While neural implicit functions have previously been applied to this task, their application to mapping anatomical consistency across subjects has been limited. In this work, we introduce Neural Implicit Heart Coordinates (NIHCs), a standardized implicit coordinate system, based on universal ventricular coordinates, that provides a common anatomical reference frame for the human heart. Our method predicts NIHCs directly from a limited number of 2D segmentations (sparse acquisition) and subsequently decodes them into dense 3D segmentations and high-resolution meshes at arbitrary output resolution. Trained on a large dataset of 5,000 cardiac meshes, the model achieves high reconstruction accuracy on clinical contours, with mean Euclidean surface errors of 2.51$\pm$0.33 mm in a diseased cohort (n=4549) and 2.3$\pm$0.36 mm in a healthy cohort (n=5576). The NIHC representation enables anatomically coherent reconstruction even under severe slice sparsity and segmentation noise, faithfully recovering complex structures such as the valve planes. Compared with traditional pipelines, inference time is reduced from over 60 s to 5-15 s. These results demonstrate that NIHCs constitute a robust and efficient anatomical representation for patient-specific 3D cardiac reconstruction from minimal input data.
Debiasing Cardiac Imaging with Controlled Latent Diffusion Models
Mar 28, 2024

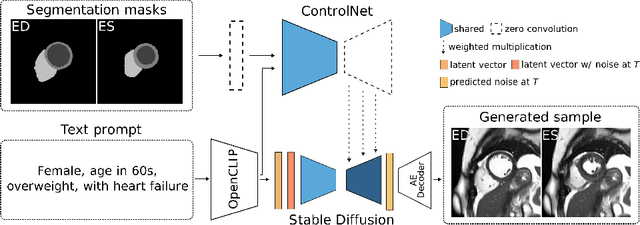

Abstract:The progress in deep learning solutions for disease diagnosis and prognosis based on cardiac magnetic resonance imaging is hindered by highly imbalanced and biased training data. To address this issue, we propose a method to alleviate imbalances inherent in datasets through the generation of synthetic data based on sensitive attributes such as sex, age, body mass index, and health condition. We adopt ControlNet based on a denoising diffusion probabilistic model to condition on text assembled from patient metadata and cardiac geometry derived from segmentation masks using a large-cohort study, specifically, the UK Biobank. We assess our method by evaluating the realism of the generated images using established quantitative metrics. Furthermore, we conduct a downstream classification task aimed at debiasing a classifier by rectifying imbalances within underrepresented groups through synthetically generated samples. Our experiments demonstrate the effectiveness of the proposed approach in mitigating dataset imbalances, such as the scarcity of younger patients or individuals with normal BMI level suffering from heart failure. This work represents a major step towards the adoption of synthetic data for the development of fair and generalizable models for medical classification tasks. Notably, we conduct all our experiments using a single, consumer-level GPU to highlight the feasibility of our approach within resource-constrained environments. Our code is available at https://github.com/faildeny/debiasing-cardiac-mri.
Fully Automated Myocardial Strain Estimation from CMR Tagged Images using a Deep Learning Framework in the UK Biobank
Apr 15, 2020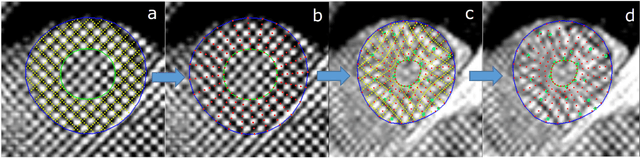
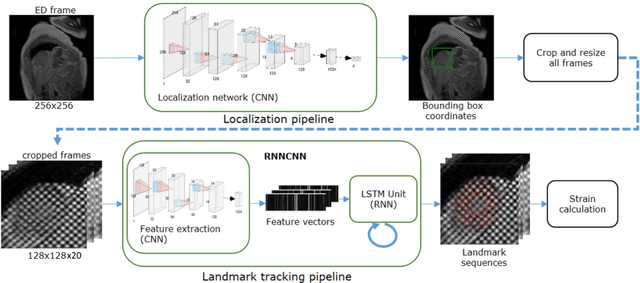
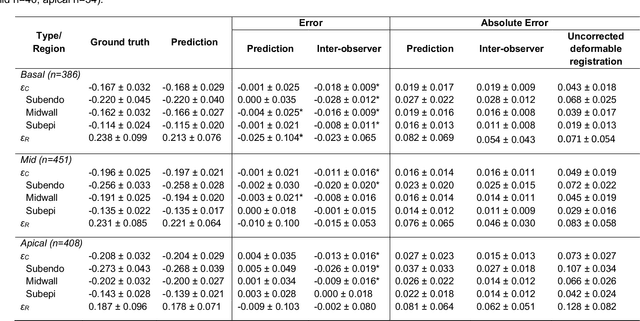
Abstract:Purpose: To demonstrate the feasibility and performance of a fully automated deep learning framework to estimate myocardial strain from short-axis cardiac magnetic resonance tagged images. Methods and Materials: In this retrospective cross-sectional study, 4508 cases from the UK Biobank were split randomly into 3244 training and 812 validation cases, and 452 test cases. Ground truth myocardial landmarks were defined and tracked by manual initialization and correction of deformable image registration using previously validated software with five readers. The fully automatic framework consisted of 1) a convolutional neural network (CNN) for localization, and 2) a combination of a recurrent neural network (RNN) and a CNN to detect and track the myocardial landmarks through the image sequence for each slice. Radial and circumferential strain were then calculated from the motion of the landmarks and averaged on a slice basis. Results: Within the test set, myocardial end-systolic circumferential Green strain errors were -0.001 +/- 0.025, -0.001 +/- 0.021, and 0.004 +/- 0.035 in basal, mid, and apical slices respectively (mean +/- std. dev. of differences between predicted and manual strain). The framework reproduced significant reductions in circumferential strain in diabetics, hypertensives, and participants with previous heart attack. Typical processing time was ~260 frames (~13 slices) per second on an NVIDIA Tesla K40 with 12GB RAM, compared with 6-8 minutes per slice for the manual analysis. Conclusions: The fully automated RNNCNN framework for analysis of myocardial strain enabled unbiased strain evaluation in a high-throughput workflow, with similar ability to distinguish impairment due to diabetes, hypertension, and previous heart attack.
* accepted in Radiology Cardiothoracic Imaging
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge