Alexis Allot
for the AREDS2 Deep Learning Research Group
PubTator 3.0: an AI-powered Literature Resource for Unlocking Biomedical Knowledge
Jan 19, 2024

Abstract:PubTator 3.0 (https://www.ncbi.nlm.nih.gov/research/pubtator3/) is a biomedical literature resource using state-of-the-art AI techniques to offer semantic and relation searches for key concepts like proteins, genetic variants, diseases, and chemicals. It currently provides over one billion entity and relation annotations across approximately 36 million PubMed abstracts and 6 million full-text articles from the PMC open access subset, updated weekly. PubTator 3.0's online interface and API utilize these precomputed entity relations and synonyms to provide advanced search capabilities and enable large-scale analyses, streamlining many complex information needs. We showcase the retrieval quality of PubTator 3.0 using a series of entity pair queries, demonstrating that PubTator 3.0 retrieves a greater number of articles than either PubMed or Google Scholar, with higher precision in the top 20 results. We further show that integrating ChatGPT (GPT-4) with PubTator APIs dramatically improves the factuality and verifiability of its responses. In summary, PubTator 3.0 offers a comprehensive set of features and tools that allow researchers to navigate the ever-expanding wealth of biomedical literature, expediting research and unlocking valuable insights for scientific discovery.
LitCovid in 2022: an information resource for the COVID-19 literature
Sep 27, 2022
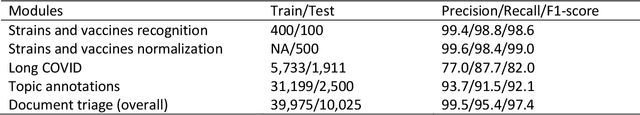

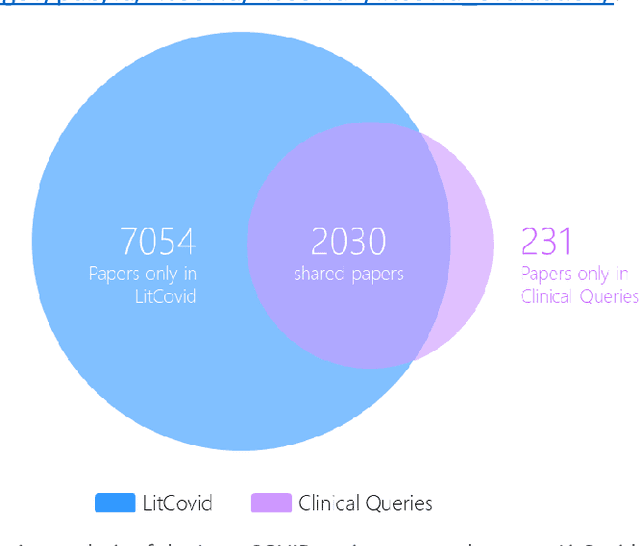
Abstract:LitCovid (https://www.ncbi.nlm.nih.gov/research/coronavirus/), first launched in February 2020, is a first-of-its-kind literature hub for tracking up-to-date published research on COVID-19. The number of articles in LitCovid has increased from 55,000 to ~300,000 over the past two and half years, with a consistent growth rate of ~10,000 articles per month. In addition to the rapid literature growth, the COVID-19 pandemic has evolved dramatically. For instance, the Omicron variant has now accounted for over 98% of new infections in the U.S. In response to the continuing evolution of the COVID-19 pandemic, this article describes significant updates to LitCovid over the last two years. First, we introduced the Long Covid collection consisting of the articles on COVID-19 survivors experiencing ongoing multisystemic symptoms, including respiratory issues, cardiovascular disease, cognitive impairment, and profound fatigue. Second, we provided new annotations on the latest COVID-19 strains and vaccines mentioned in the literature. Third, we improved several existing features with more accurate machine learning algorithms for annotating topics and classifying articles relevant to COVID-19. LitCovid has been widely used with millions of accesses by users worldwide on various information needs and continues to play a critical role in collecting, curating, and standardizing the latest knowledge on the COVID-19 literature.
Comprehensive identification of Long Covid articles with human-in-the-loop machine learning
Sep 16, 2022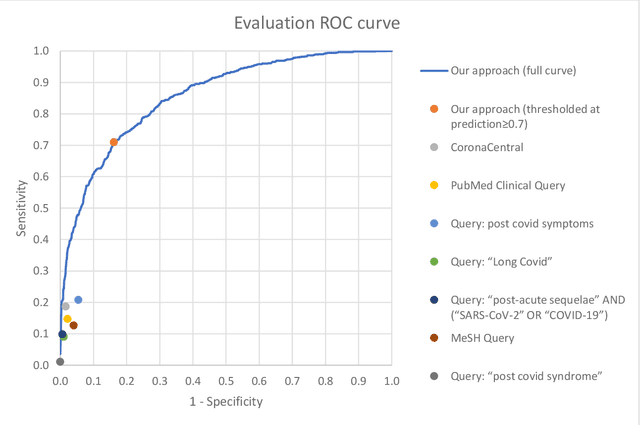
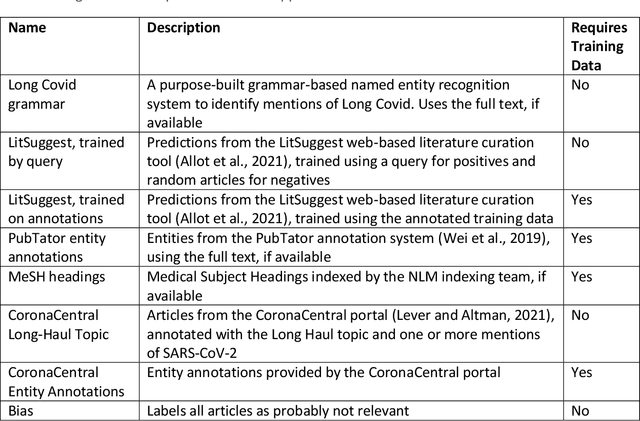
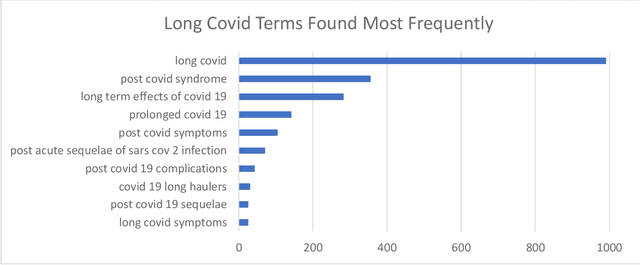
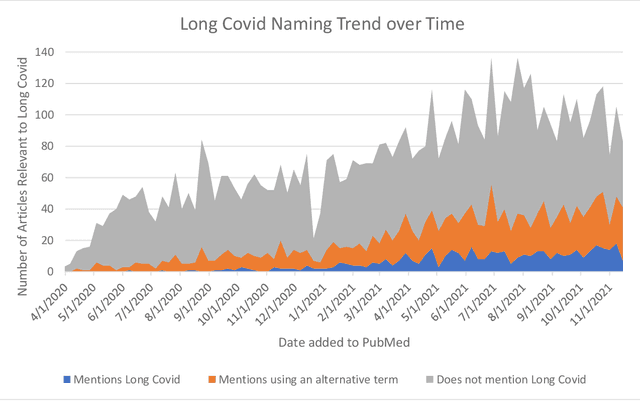
Abstract:A significant percentage of COVID-19 survivors experience ongoing multisystemic symptoms that often affect daily living, a condition known as Long Covid or post-acute-sequelae of SARS-CoV-2 infection. However, identifying Long Covid articles is challenging since articles refer to the condition using a variety of less common terms or refrain from naming it at all. We developed an iterative human-in-the-loop machine learning framework designed to effectively leverage the data available and make the most efficient use of human labels. Specifically, our approach combines data programming with active learning into a robust ensemble model. Evaluating our model on a holdout set demonstrates over three times the sensitivity of other methods. We apply our model to PubMed to create the Long Covid collection, and demonstrate that (1) most Long Covid articles do not refer to Long Covid by any name (2) when the condition is named, the name used most frequently in the biomedical literature is Long Covid, and (3) Long Covid is associated with disorders in a wide variety of body systems. The Long Covid collection is updated weekly and is searchable online at the LitCovid portal: https://www.ncbi.nlm.nih.gov/research/coronavirus/docsum?filters=e_condition.LongCovid
Multi-label classification for biomedical literature: an overview of the BioCreative VII LitCovid Track for COVID-19 literature topic annotations
Apr 20, 2022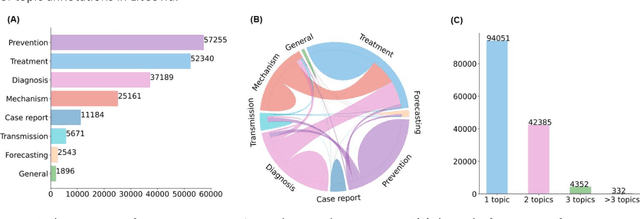
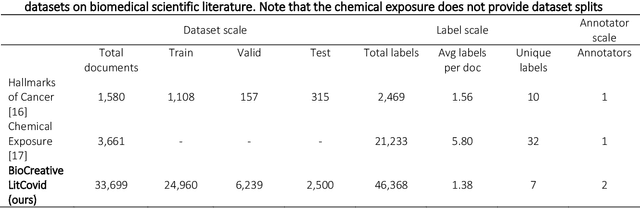
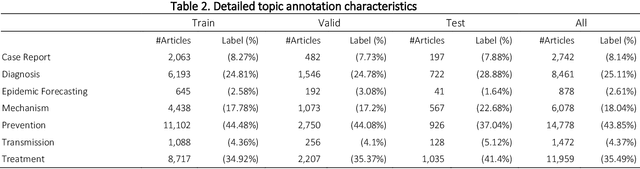
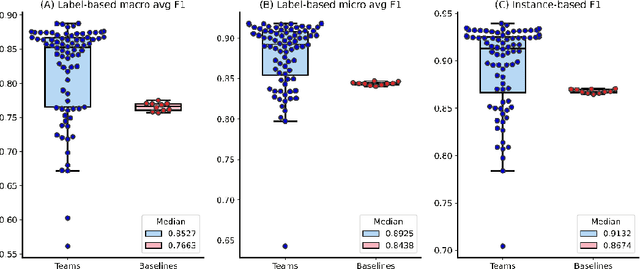
Abstract:The COVID-19 pandemic has been severely impacting global society since December 2019. Massive research has been undertaken to understand the characteristics of the virus and design vaccines and drugs. The related findings have been reported in biomedical literature at a rate of about 10,000 articles on COVID-19 per month. Such rapid growth significantly challenges manual curation and interpretation. For instance, LitCovid is a literature database of COVID-19-related articles in PubMed, which has accumulated more than 200,000 articles with millions of accesses each month by users worldwide. One primary curation task is to assign up to eight topics (e.g., Diagnosis and Treatment) to the articles in LitCovid. Despite the continuing advances in biomedical text mining methods, few have been dedicated to topic annotations in COVID-19 literature. To close the gap, we organized the BioCreative LitCovid track to call for a community effort to tackle automated topic annotation for COVID-19 literature. The BioCreative LitCovid dataset, consisting of over 30,000 articles with manually reviewed topics, was created for training and testing. It is one of the largest multilabel classification datasets in biomedical scientific literature. 19 teams worldwide participated and made 80 submissions in total. Most teams used hybrid systems based on transformers. The highest performing submissions achieved 0.8875, 0.9181, and 0.9394 for macro F1-score, micro F1-score, and instance-based F1-score, respectively. The level of participation and results demonstrate a successful track and help close the gap between dataset curation and method development. The dataset is publicly available via https://ftp.ncbi.nlm.nih.gov/pub/lu/LitCovid/biocreative/ for benchmarking and further development.
LitMC-BERT: transformer-based multi-label classification of biomedical literature with an application on COVID-19 literature curation
Apr 19, 2022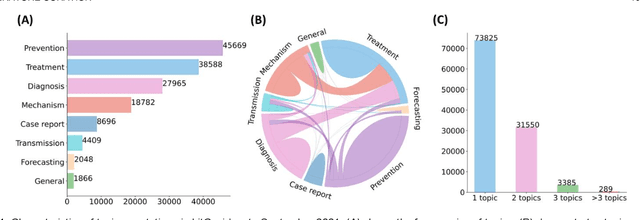


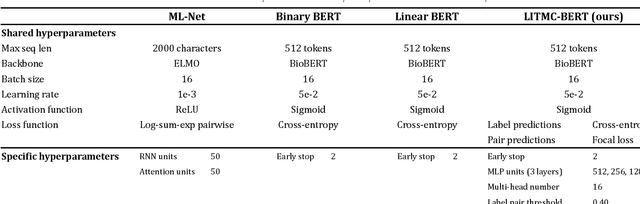
Abstract:The rapid growth of biomedical literature poses a significant challenge for curation and interpretation. This has become more evident during the COVID-19 pandemic. LitCovid, a literature database of COVID-19 related papers in PubMed, has accumulated over 180,000 articles with millions of accesses. Approximately 10,000 new articles are added to LitCovid every month. A main curation task in LitCovid is topic annotation where an article is assigned with up to eight topics, e.g., Treatment and Diagnosis. The annotated topics have been widely used both in LitCovid (e.g., accounting for ~18% of total uses) and downstream studies such as network generation. However, it has been a primary curation bottleneck due to the nature of the task and the rapid literature growth. This study proposes LITMC-BERT, a transformer-based multi-label classification method in biomedical literature. It uses a shared transformer backbone for all the labels while also captures label-specific features and the correlations between label pairs. We compare LITMC-BERT with three baseline models on two datasets. Its micro-F1 and instance-based F1 are 5% and 4% higher than the current best results, respectively, and only requires ~18% of the inference time than the Binary BERT baseline. The related datasets and models are available via https://github.com/ncbi/ml-transformer.
tmVar 3.0: an improved variant concept recognition and normalization tool
Apr 07, 2022

Abstract:Previous studies have shown that automated text-mining tools are becoming increasingly important for successfully unlocking variant information in scientific literature at large scale. Despite multiple attempts in the past, existing tools are still of limited recognition scope and precision. We propose tmVar 3.0: an improved variant recognition and normalization tool. Compared to its predecessors, tmVar 3.0 is able to recognize a wide spectrum of variant related entities (e.g., allele and copy number variants), and to group different variant mentions belonging to the same concept in an article for improved accuracy. Moreover, tmVar3 provides additional variant normalization options such as allele-specific identifiers from the ClinGen Allele Registry. tmVar3 exhibits a state-of-the-art performance with over 90% accuracy in F-measure in variant recognition and normalization, when evaluated on three independent benchmarking datasets. tmVar3 is freely available for download. We have also processed the entire PubMed and PMC with tmVar3 and released its annotations on our FTP. Availability: ftp://ftp.ncbi.nlm.nih.gov/pub/lu/tmVar3
Multi-modal, multi-task, multi-attention deep learning detection of reticular pseudodrusen: towards automated and accessible classification of age-related macular degeneration
Nov 11, 2020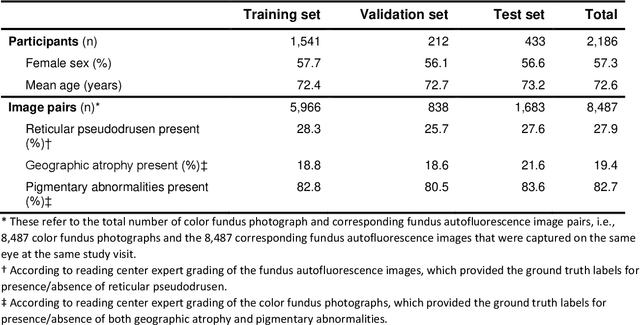
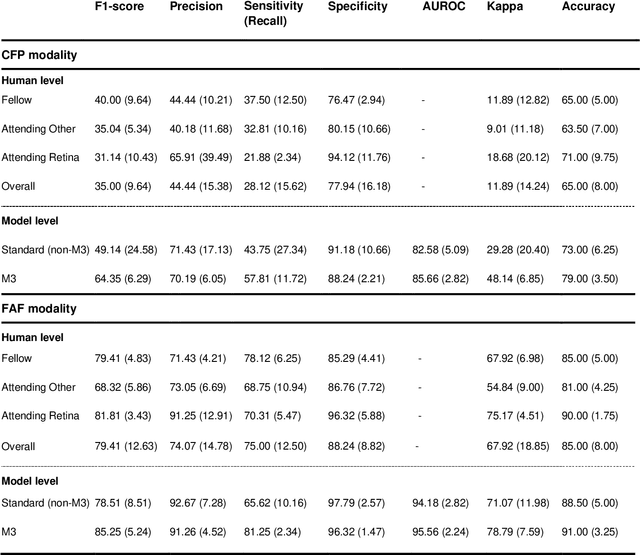
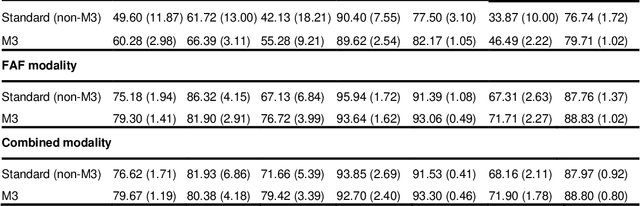
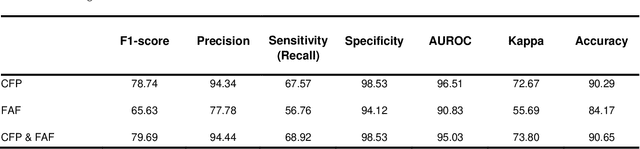
Abstract:Objective Reticular pseudodrusen (RPD), a key feature of age-related macular degeneration (AMD), are poorly detected by human experts on standard color fundus photography (CFP) and typically require advanced imaging modalities such as fundus autofluorescence (FAF). The objective was to develop and evaluate the performance of a novel 'M3' deep learning framework on RPD detection. Materials and Methods A deep learning framework M3 was developed to detect RPD presence accurately using CFP alone, FAF alone, or both, employing >8000 CFP-FAF image pairs obtained prospectively (Age-Related Eye Disease Study 2). The M3 framework includes multi-modal (detection from single or multiple image modalities), multi-task (training different tasks simultaneously to improve generalizability), and multi-attention (improving ensembled feature representation) operation. Performance on RPD detection was compared with state-of-the-art deep learning models and 13 ophthalmologists; performance on detection of two other AMD features (geographic atrophy and pigmentary abnormalities) was also evaluated. Results For RPD detection, M3 achieved area under receiver operating characteristic (AUROC) 0.832, 0.931, and 0.933 for CFP alone, FAF alone, and both, respectively. M3 performance on CFP was very substantially superior to human retinal specialists (median F1-score 0.644 versus 0.350). External validation (on Rotterdam Study, Netherlands) demonstrated high accuracy on CFP alone (AUROC 0.965). The M3 framework also accurately detected geographic atrophy and pigmentary abnormalities (AUROC 0.909 and 0.912, respectively), demonstrating its generalizability. Conclusion This study demonstrates the successful development, robust evaluation, and external validation of a novel deep learning framework that enables accessible, accurate, and automated AMD diagnosis and prognosis.
Navigating the landscape of COVID-19 research through literature analysis: A bird's eye view
Sep 11, 2020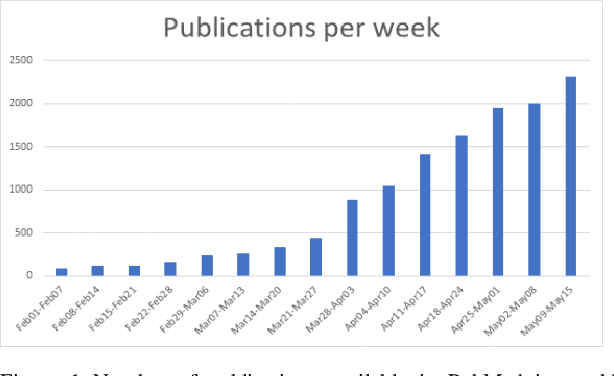
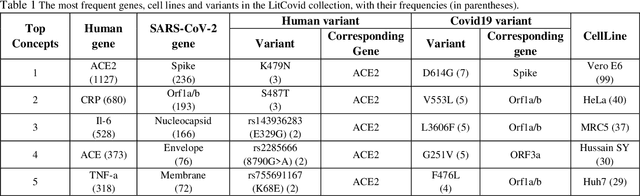
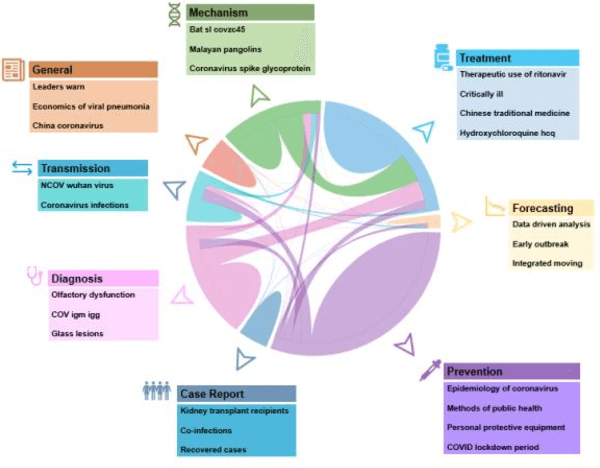
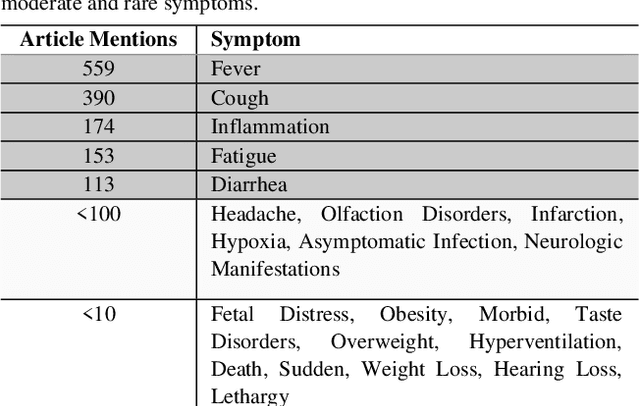
Abstract:Timely access to accurate scientific literature in the battle with the ongoing COVID-19 pandemic is critical. This unprecedented public health risk has motivated research towards understanding the disease in general, identifying drugs to treat the disease, developing potential vaccines, etc. This has given rise to a rapidly growing body of literature that doubles in number of publications every 20 days as of May 2020. Providing medical professionals with means to quickly analyze the literature and discover growing areas of knowledge is necessary for addressing their question and information needs. In this study we analyze the LitCovid collection, 13,369 COVID-19 related articles found in PubMed as of May 15th, 2020 with the purpose of examining the landscape of literature and presenting it in a format that facilitates information navigation and understanding. We do that by applying state-of-the-art named entity recognition, classification, clustering and other NLP techniques. By applying NER tools, we capture relevant bioentities (such as diseases, internal body organs, etc.) and assess the strength of their relationship with COVID-19 by the extent they are discussed in the corpus. We also collect a variety of symptoms and co-morbidities discussed in reference to COVID-19. Our clustering algorithm identifies topics represented by groups of related terms, and computes clusters corresponding to documents associated with the topic terms. Among the topics we observe several that persist through the duration of multiple weeks and have numerous associated documents, as well several that appear as emerging topics with fewer documents. All the tools and data are publicly available, and this framework can be applied to any literature collection. Taken together, these analyses produce a comprehensive, synthesized view of COVID-19 research to facilitate knowledge discovery from literature.
* 10 pages, 8 Figures, Submitted to KDD 2020 Health Day
Predicting risk of late age-related macular degeneration using deep learning
Jul 19, 2020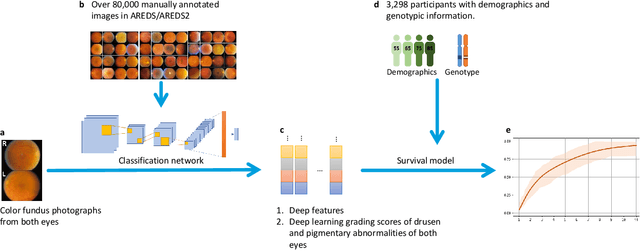
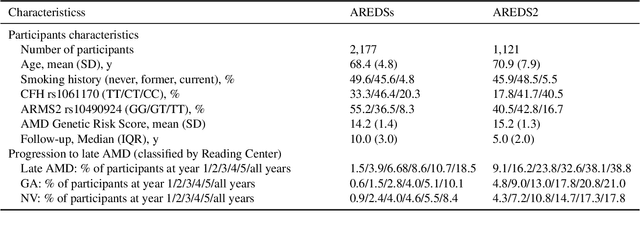
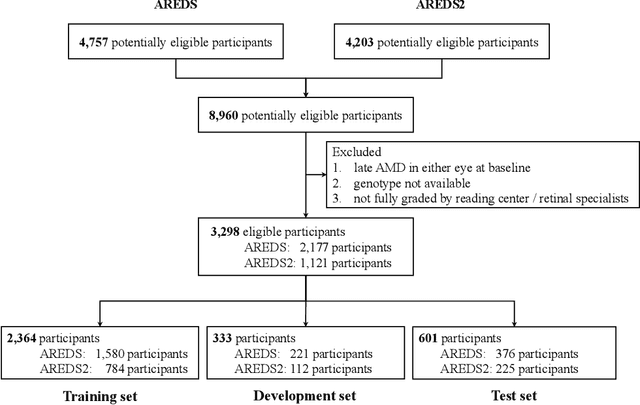
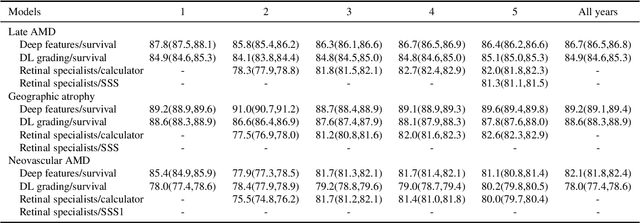
Abstract:By 2040, age-related macular degeneration (AMD) will affect approximately 288 million people worldwide. Identifying individuals at high risk of progression to late AMD, the sight-threatening stage, is critical for clinical actions, including medical interventions and timely monitoring. Although deep learning has shown promise in diagnosing/screening AMD using color fundus photographs, it remains difficult to predict individuals' risks of late AMD accurately. For both tasks, these initial deep learning attempts have remained largely unvalidated in independent cohorts. Here, we demonstrate how deep learning and survival analysis can predict the probability of progression to late AMD using 3,298 participants (over 80,000 images) from the Age-Related Eye Disease Studies AREDS and AREDS2, the largest longitudinal clinical trials in AMD. When validated against an independent test dataset of 601 participants, our model achieved high prognostic accuracy (five-year C-statistic 86.4 (95% confidence interval 86.2-86.6)) that substantially exceeded that of retinal specialists using two existing clinical standards (81.3 (81.1-81.5) and 82.0 (81.8-82.3), respectively). Interestingly, our approach offers additional strengths over the existing clinical standards in AMD prognosis (e.g., risk ascertainment above 50%) and is likely to be highly generalizable, given the breadth of training data from 82 US retinal specialty clinics. Indeed, during external validation through training on AREDS and testing on AREDS2 as an independent cohort, our model retained substantially higher prognostic accuracy than existing clinical standards. These results highlight the potential of deep learning systems to enhance clinical decision-making in AMD patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge