Li Fang
AI-enabled Satellite Edge Computing: A Single-Pixel Feature based Shallow Classification Model for Hyperspectral Imaging
Jan 26, 2026Abstract:As the important component of the Earth observation system, hyperspectral imaging satellites provide high-fidelity and enriched information for the formulation of related policies due to the powerful spectral measurement capabilities. However, the transmission speed of the satellite downlink has become a major bottleneck in certain applications, such as disaster monitoring and emergency mapping, which demand a fast response ability. We propose an efficient AI-enabled Satellite Edge Computing paradigm for hyperspectral image classification, facilitating the satellites to attain autonomous decision-making. To accommodate the resource constraints of satellite platforms, the proposed method adopts a lightweight, non-deep learning framework integrated with a few-shot learning strategy. Moreover, onboard processing on satellites could be faced with sensor failure and scan pattern errors, which result in degraded image quality with bad/misaligned pixels and mixed noise. To address these challenges, we develop a novel two-stage pixel-wise label propagation scheme that utilizes only intrinsic spectral features at the single pixel level without the necessity to consider spatial structural information as requested by deep neural networks. In the first stage, initial pixel labels are obtained by propagating selected anchor labels through the constructed anchor-pixel affinity matrix. Subsequently, a top-k pruned sparse graph is generated by directly computing pixel-level similarities. In the second stage, a closed-form solution derived from the sparse graph is employed to replace iterative computations. Furthermore, we developed a rank constraint-based graph clustering algorithm to determine the anchor labels.
Cross-Domain Transfer with Self-Supervised Spectral-Spatial Modeling for Hyperspectral Image Classification
Jan 26, 2026Abstract:Self-supervised learning has demonstrated considerable potential in hyperspectral representation, yet its application in cross-domain transfer scenarios remains under-explored. Existing methods, however, still rely on source domain annotations and are susceptible to distribution shifts, leading to degraded generalization performance in the target domain. To address this, this paper proposes a self-supervised cross-domain transfer framework that learns transferable spectral-spatial joint representations without source labels and achieves efficient adaptation under few samples in the target domain. During the self-supervised pre-training phase, a Spatial-Spectral Transformer (S2Former) module is designed. It adopts a dual-branch spatial-spectral transformer and introduces a bidirectional cross-attention mechanism to achieve spectral-spatial collaborative modeling: the spatial branch enhances structural awareness through random masking, while the spectral branch captures fine-grained differences. Both branches mutually guide each other to improve semantic consistency. We further propose a Frequency Domain Constraint (FDC) to maintain frequency-domain consistency through real Fast Fourier Transform (rFFT) and high-frequency magnitude loss, thereby enhancing the model's capability to discern fine details and boundaries. During the fine-tuning phase, we introduce a Diffusion-Aligned Fine-tuning (DAFT) distillation mechanism. This aligns semantic evolution trajectories through a teacher-student structure, enabling robust transfer learning under low-label conditions. Experimental results demonstrate stable classification performance and strong cross-domain adaptability across four hyperspectral datasets, validating the method's effectiveness under resource-constrained conditions.
Depth-Guided Bundle Sampling for Efficient Generalizable Neural Radiance Field Reconstruction
May 26, 2025Abstract:Recent advancements in generalizable novel view synthesis have achieved impressive quality through interpolation between nearby views. However, rendering high-resolution images remains computationally intensive due to the need for dense sampling of all rays. Recognizing that natural scenes are typically piecewise smooth and sampling all rays is often redundant, we propose a novel depth-guided bundle sampling strategy to accelerate rendering. By grouping adjacent rays into a bundle and sampling them collectively, a shared representation is generated for decoding all rays within the bundle. To further optimize efficiency, our adaptive sampling strategy dynamically allocates samples based on depth confidence, concentrating more samples in complex regions while reducing them in smoother areas. When applied to ENeRF, our method achieves up to a 1.27 dB PSNR improvement and a 47% increase in FPS on the DTU dataset. Extensive experiments on synthetic and real-world datasets demonstrate state-of-the-art rendering quality and up to 2x faster rendering compared to existing generalizable methods. Code is available at https://github.com/KLMAV-CUC/GDB-NeRF.
GoLF-NRT: Integrating Global Context and Local Geometry for Few-Shot View Synthesis
May 26, 2025Abstract:Neural Radiance Fields (NeRF) have transformed novel view synthesis by modeling scene-specific volumetric representations directly from images. While generalizable NeRF models can generate novel views across unknown scenes by learning latent ray representations, their performance heavily depends on a large number of multi-view observations. However, with limited input views, these methods experience significant degradation in rendering quality. To address this limitation, we propose GoLF-NRT: a Global and Local feature Fusion-based Neural Rendering Transformer. GoLF-NRT enhances generalizable neural rendering from few input views by leveraging a 3D transformer with efficient sparse attention to capture global scene context. In parallel, it integrates local geometric features extracted along the epipolar line, enabling high-quality scene reconstruction from as few as 1 to 3 input views. Furthermore, we introduce an adaptive sampling strategy based on attention weights and kernel regression, improving the accuracy of transformer-based neural rendering. Extensive experiments on public datasets show that GoLF-NRT achieves state-of-the-art performance across varying numbers of input views, highlighting the effectiveness and superiority of our approach. Code is available at https://github.com/KLMAV-CUC/GoLF-NRT.
Enhancing Features in Long-tailed Data Using Large Vision Mode
Apr 15, 2025Abstract:Language-based foundation models, such as large language models (LLMs) or large vision-language models (LVLMs), have been widely studied in long-tailed recognition. However, the need for linguistic data is not applicable to all practical tasks. In this study, we aim to explore using large vision models (LVMs) or visual foundation models (VFMs) to enhance long-tailed data features without any language information. Specifically, we extract features from the LVM and fuse them with features in the baseline network's map and latent space to obtain the augmented features. Moreover, we design several prototype-based losses in the latent space to further exploit the potential of the augmented features. In the experimental section, we validate our approach on two benchmark datasets: ImageNet-LT and iNaturalist2018.
Exploiting Large Language Models Capabilities for Question Answer-Driven Knowledge Graph Completion Across Static and Temporal Domains
Aug 20, 2024



Abstract:Knowledge graph completion (KGC) aims to identify missing triples in a knowledge graph (KG). This is typically achieved through tasks such as link prediction and instance completion. However, these methods often focus on either static knowledge graphs (SKGs) or temporal knowledge graphs (TKGs), addressing only within-scope triples. This paper introduces a new generative completion framework called Generative Subgraph-based KGC (GS-KGC). GS-KGC employs a question-answering format to directly generate target entities, addressing the challenge of questions having multiple possible answers. We propose a strategy that extracts subgraphs centered on entities and relationships within the KG, from which negative samples and neighborhood information are separately obtained to address the one-to-many problem. Our method generates negative samples using known facts to facilitate the discovery of new information. Furthermore, we collect and refine neighborhood path data of known entities, providing contextual information to enhance reasoning in large language models (LLMs). Our experiments evaluated the proposed method on four SKGs and two TKGs, achieving state-of-the-art Hits@1 metrics on five datasets. Analysis of the results shows that GS-KGC can discover new triples within existing KGs and generate new facts beyond the closed KG, effectively bridging the gap between closed-world and open-world KGC.
A Radiometric Correction based Optical Modeling Approach to Removing Reflection Noise in TLS Point Clouds of Urban Scenes
Jul 03, 2024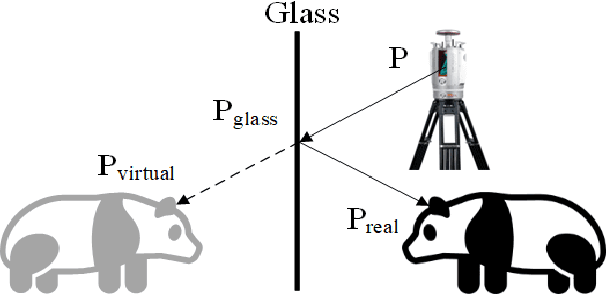
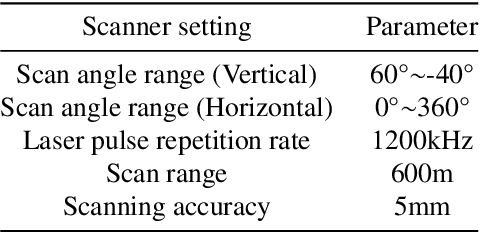
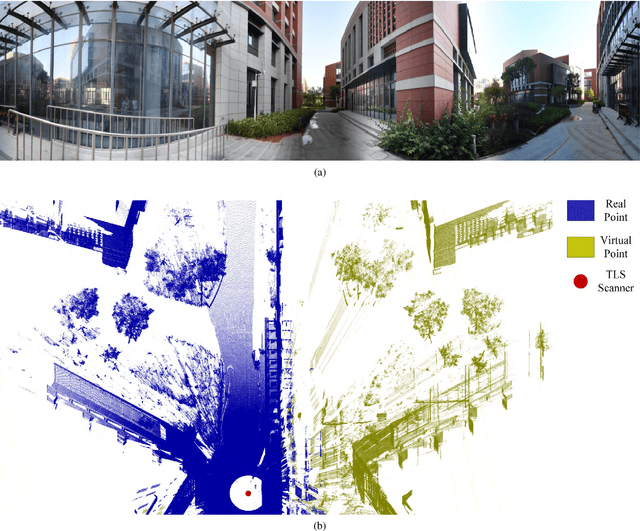
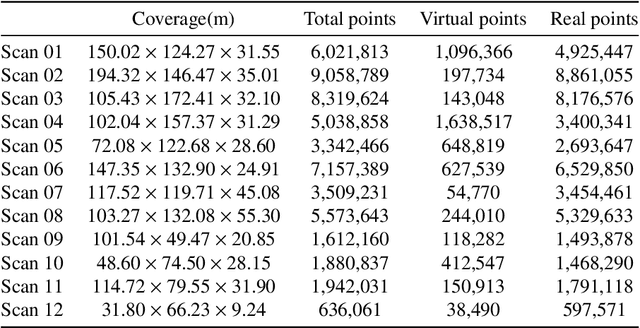
Abstract:Point clouds are vital in computer vision tasks such as 3D reconstruction, autonomous driving, and robotics. However, TLS-acquired point clouds often contain virtual points from reflective surfaces, causing disruptions. This study presents a reflection noise elimination algorithm for TLS point clouds. Our innovative reflection plane detection algorithm, based on geometry-optical models and physical properties, identifies and categorizes reflection points per optical reflection theory. We've adapted the LSFH feature descriptor to retain reflection features, mitigating interference from symmetrical architectural structures. By incorporating the Hausdorff feature distance, the algorithm enhances resilience to ghosting and deformation, improving virtual point detection accuracy. Extensive experiments on the 3DRN benchmark dataset, featuring diverse urban environments with virtual TLS reflection noise, show our algorithm improves precision and recall rates for 3D points in reflective regions by 57.03\% and 31.80\%, respectively. Our method achieves a 9.17\% better outlier detection rate and 5.65\% higher accuracy than leading methods. Access the 3DRN dataset at (https://github.com/Tsuiky/3DRN).
Shape-programmable Adaptive Multi-material Microrobots for Biomedical Applications
Dec 31, 2023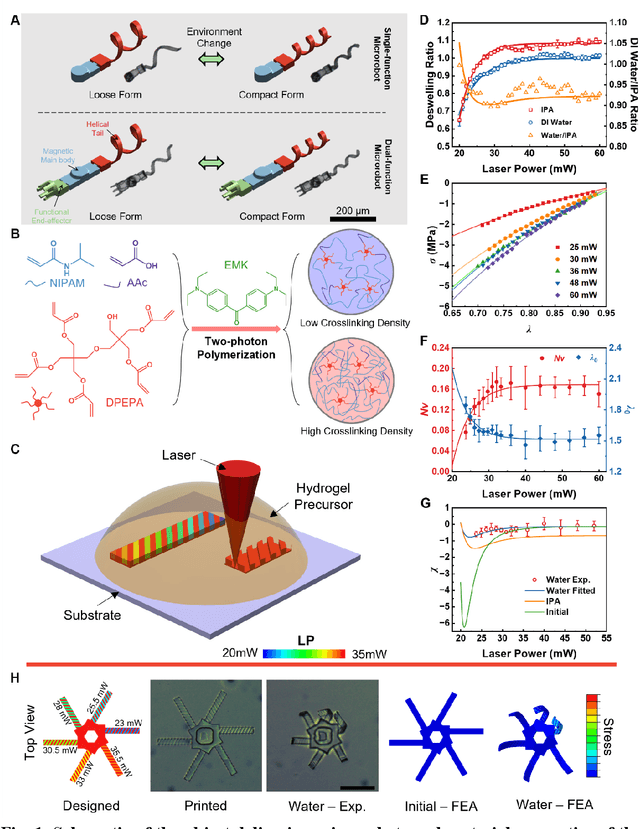
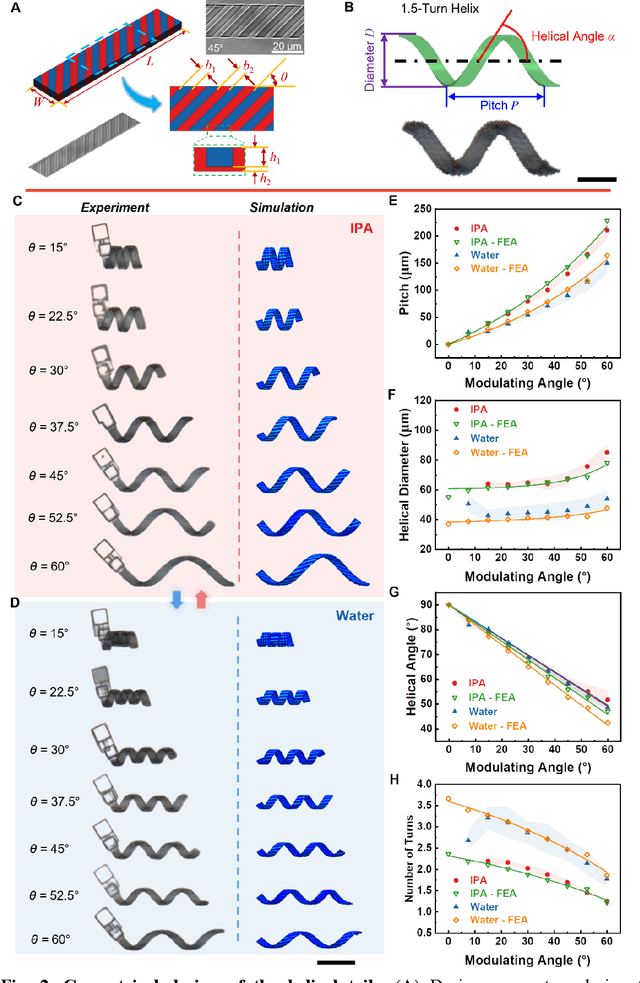
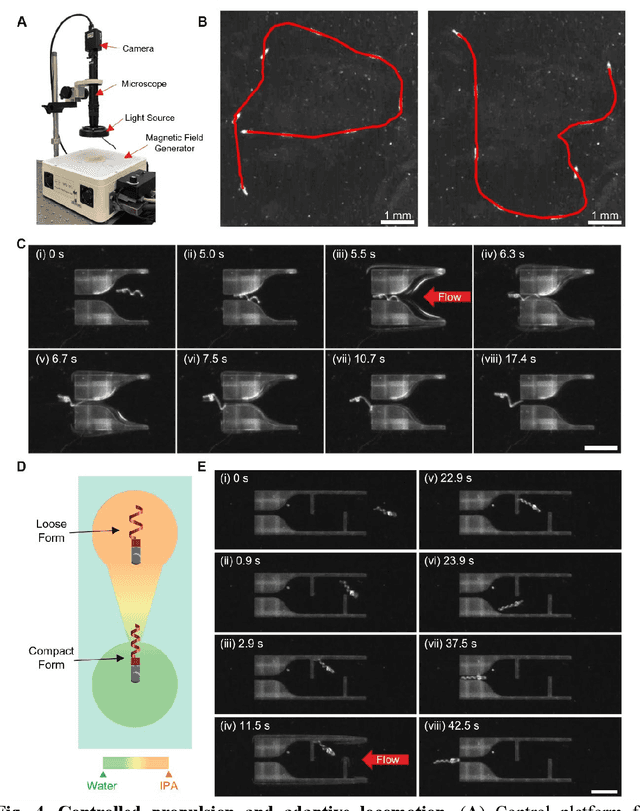
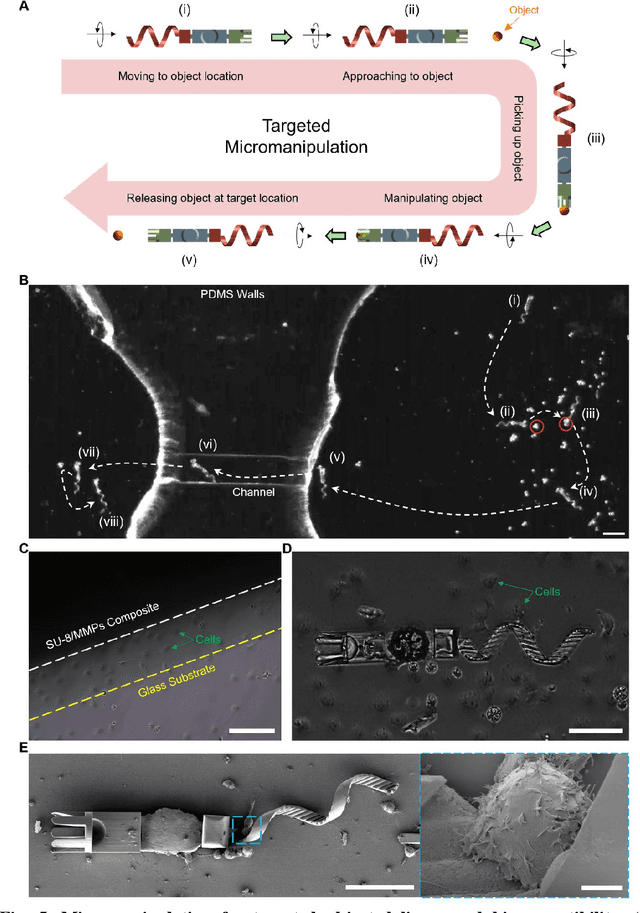
Abstract:Flagellated microorganisms can swim at low Reynolds numbers and adapt to changes in their environment. Specifically, the flagella can switch their shapes or modes through gene expression. In the past decade, efforts have been made to fabricate and investigate rigid types of microrobots without any adaptation to the environments. More recently, obtaining adaptive microrobots mimicking real microorganisms is getting more attention. However, even though some adaptive microrobots achieved by hydrogels have emerged, the swimming behaviors of the microrobots before and after the environment-induced deformations are not predicted in a systematic standardized way. In this work, experiments, finite element analysis, and dynamic modeling are presented together to realize a complete understanding of these adaptive microrobots. The above three parts are cross-verified proving the success of using such methods, facilitating the bio-applications with shape-programmable and even swimming performance-programmable microrobots. Moreover, an application of targeted object delivery using the proposed microrobot has been successfully demonstrated. Finally, cytotoxicity tests are performed to prove the potential for using the proposed microrobot for biomedical applications.
CP-KGC: Constrained-Prompt Knowledge Graph Completion with Large Language Models
Oct 12, 2023



Abstract:Knowledge graph completion (KGC) aims to utilize existing knowledge to deduce and infer missing connections within knowledge graphs. Text-based approaches, like SimKGC, have outperformed graph embedding methods, showcasing the promise of inductive KGC. However, the efficacy of text-based methods hinges on the quality of entity textual descriptions. In this paper, we identify the key issue of whether large language models (LLMs) can generate effective text. To mitigate hallucination in LLM-generated text in this paper, we introduce a constraint-based prompt that utilizes the entity and its textual description as contextual constraints to enhance data quality. Our Constrained-Prompt Knowledge Graph Completion (CP-KGC) method demonstrates effective inference under low resource computing conditions and surpasses prior results on the WN18RR and FB15K237 datasets. This showcases the integration of LLMs in KGC tasks and provides new directions for future research.
Bioformer: an efficient transformer language model for biomedical text mining
Feb 03, 2023Abstract:Pretrained language models such as Bidirectional Encoder Representations from Transformers (BERT) have achieved state-of-the-art performance in natural language processing (NLP) tasks. Recently, BERT has been adapted to the biomedical domain. Despite the effectiveness, these models have hundreds of millions of parameters and are computationally expensive when applied to large-scale NLP applications. We hypothesized that the number of parameters of the original BERT can be dramatically reduced with minor impact on performance. In this study, we present Bioformer, a compact BERT model for biomedical text mining. We pretrained two Bioformer models (named Bioformer8L and Bioformer16L) which reduced the model size by 60% compared to BERTBase. Bioformer uses a biomedical vocabulary and was pre-trained from scratch on PubMed abstracts and PubMed Central full-text articles. We thoroughly evaluated the performance of Bioformer as well as existing biomedical BERT models including BioBERT and PubMedBERT on 15 benchmark datasets of four different biomedical NLP tasks: named entity recognition, relation extraction, question answering and document classification. The results show that with 60% fewer parameters, Bioformer16L is only 0.1% less accurate than PubMedBERT while Bioformer8L is 0.9% less accurate than PubMedBERT. Both Bioformer16L and Bioformer8L outperformed BioBERTBase-v1.1. In addition, Bioformer16L and Bioformer8L are 2-3 fold as fast as PubMedBERT/BioBERTBase-v1.1. Bioformer has been successfully deployed to PubTator Central providing gene annotations over 35 million PubMed abstracts and 5 million PubMed Central full-text articles. We make Bioformer publicly available via https://github.com/WGLab/bioformer, including pre-trained models, datasets, and instructions for downstream use.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge