Jingcheng Du
Large language models in biomedical natural language processing: benchmarks, baselines, and recommendations
May 10, 2023Abstract:Biomedical literature is growing rapidly, making it challenging to curate and extract knowledge manually. Biomedical natural language processing (BioNLP) techniques that can automatically extract information from biomedical literature help alleviate this burden. Recently, large Language Models (LLMs), such as GPT-3 and GPT-4, have gained significant attention for their impressive performance. However, their effectiveness in BioNLP tasks and impact on method development and downstream users remain understudied. This pilot study (1) establishes the baseline performance of GPT-3 and GPT-4 at both zero-shot and one-shot settings in eight BioNLP datasets across four applications: named entity recognition, relation extraction, multi-label document classification, and semantic similarity and reasoning, (2) examines the errors produced by the LLMs and categorized the errors into three types: missingness, inconsistencies, and unwanted artificial content, and (3) provides suggestions for using LLMs in BioNLP applications. We make the datasets, baselines, and results publicly available to the community via https://github.com/qingyu-qc/gpt_bionlp_benchmark.
Multi-label classification for biomedical literature: an overview of the BioCreative VII LitCovid Track for COVID-19 literature topic annotations
Apr 20, 2022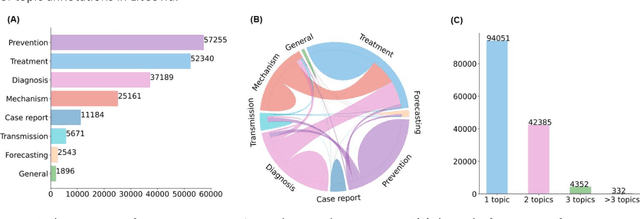
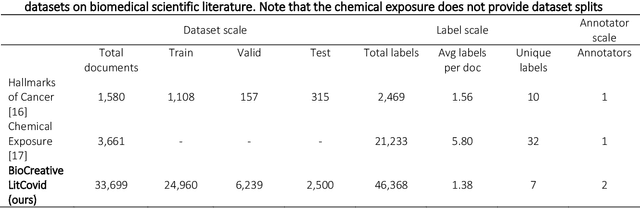
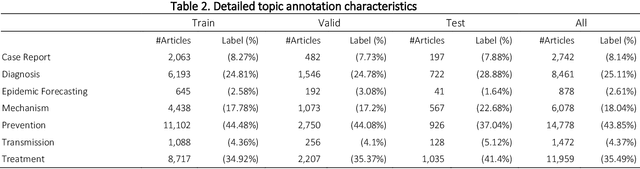
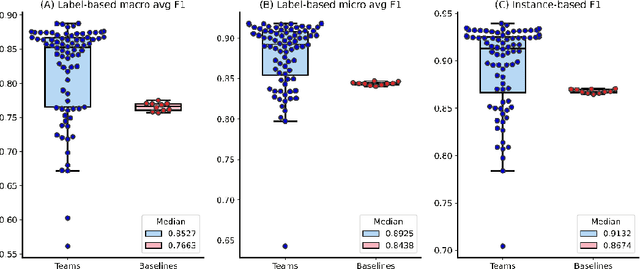
Abstract:The COVID-19 pandemic has been severely impacting global society since December 2019. Massive research has been undertaken to understand the characteristics of the virus and design vaccines and drugs. The related findings have been reported in biomedical literature at a rate of about 10,000 articles on COVID-19 per month. Such rapid growth significantly challenges manual curation and interpretation. For instance, LitCovid is a literature database of COVID-19-related articles in PubMed, which has accumulated more than 200,000 articles with millions of accesses each month by users worldwide. One primary curation task is to assign up to eight topics (e.g., Diagnosis and Treatment) to the articles in LitCovid. Despite the continuing advances in biomedical text mining methods, few have been dedicated to topic annotations in COVID-19 literature. To close the gap, we organized the BioCreative LitCovid track to call for a community effort to tackle automated topic annotation for COVID-19 literature. The BioCreative LitCovid dataset, consisting of over 30,000 articles with manually reviewed topics, was created for training and testing. It is one of the largest multilabel classification datasets in biomedical scientific literature. 19 teams worldwide participated and made 80 submissions in total. Most teams used hybrid systems based on transformers. The highest performing submissions achieved 0.8875, 0.9181, and 0.9394 for macro F1-score, micro F1-score, and instance-based F1-score, respectively. The level of participation and results demonstrate a successful track and help close the gap between dataset curation and method development. The dataset is publicly available via https://ftp.ncbi.nlm.nih.gov/pub/lu/LitCovid/biocreative/ for benchmarking and further development.
LitMC-BERT: transformer-based multi-label classification of biomedical literature with an application on COVID-19 literature curation
Apr 19, 2022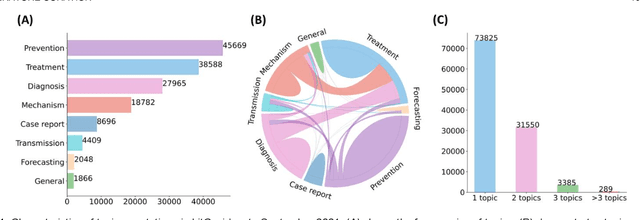


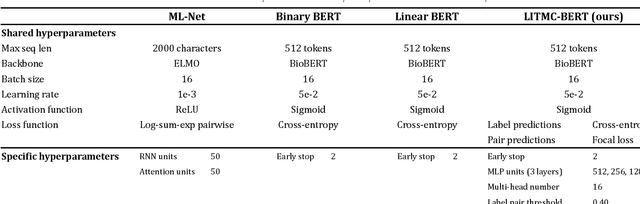
Abstract:The rapid growth of biomedical literature poses a significant challenge for curation and interpretation. This has become more evident during the COVID-19 pandemic. LitCovid, a literature database of COVID-19 related papers in PubMed, has accumulated over 180,000 articles with millions of accesses. Approximately 10,000 new articles are added to LitCovid every month. A main curation task in LitCovid is topic annotation where an article is assigned with up to eight topics, e.g., Treatment and Diagnosis. The annotated topics have been widely used both in LitCovid (e.g., accounting for ~18% of total uses) and downstream studies such as network generation. However, it has been a primary curation bottleneck due to the nature of the task and the rapid literature growth. This study proposes LITMC-BERT, a transformer-based multi-label classification method in biomedical literature. It uses a shared transformer backbone for all the labels while also captures label-specific features and the correlations between label pairs. We compare LITMC-BERT with three baseline models on two datasets. Its micro-F1 and instance-based F1 are 5% and 4% higher than the current best results, respectively, and only requires ~18% of the inference time than the Binary BERT baseline. The related datasets and models are available via https://github.com/ncbi/ml-transformer.
Mining On Alzheimer's Diseases Related Knowledge Graph to Identity Potential AD-related Semantic Triples for Drug Repurposing
Feb 17, 2022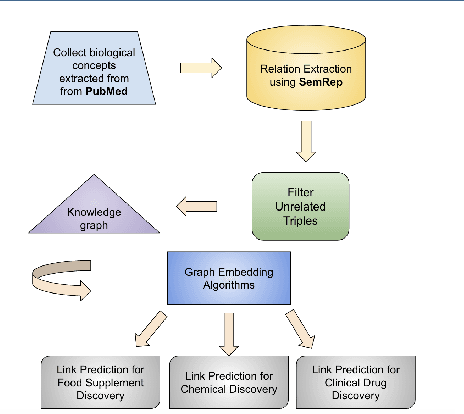

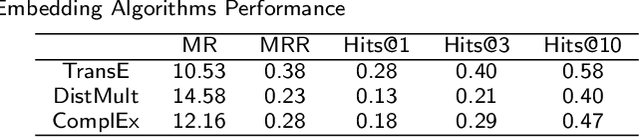
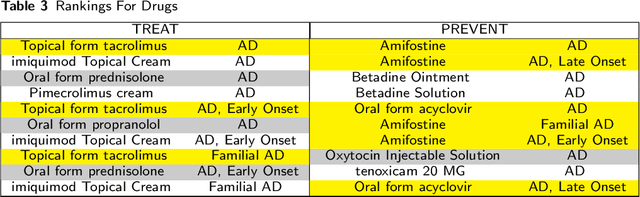
Abstract:To date, there are no effective treatments for most neurodegenerative diseases. Knowledge graphs can provide comprehensive and semantic representation for heterogeneous data, and have been successfully leveraged in many biomedical applications including drug repurposing. Our objective is to construct a knowledge graph from literature to study relations between Alzheimer's disease (AD) and chemicals, drugs and dietary supplements in order to identify opportunities to prevent or delay neurodegenerative progression. We collected biomedical annotations and extracted their relations using SemRep via SemMedDB. We used both a BERT-based classifier and rule-based methods during data preprocessing to exclude noise while preserving most AD-related semantic triples. The 1,672,110 filtered triples were used to train with knowledge graph completion algorithms (i.e., TransE, DistMult, and ComplEx) to predict candidates that might be helpful for AD treatment or prevention. Among three knowledge graph completion models, TransE outperformed the other two (MR = 13.45, Hits@1 = 0.306). We leveraged the time-slicing technique to further evaluate the prediction results. We found supporting evidence for most highly ranked candidates predicted by our model which indicates that our approach can inform reliable new knowledge. This paper shows that our graph mining model can predict reliable new relationships between AD and other entities (i.e., dietary supplements, chemicals, and drugs). The knowledge graph constructed can facilitate data-driven knowledge discoveries and the generation of novel hypotheses.
Knowledge Graph-based Neurodegenerative Diseases and Diet Relationship Discovery
Sep 13, 2021
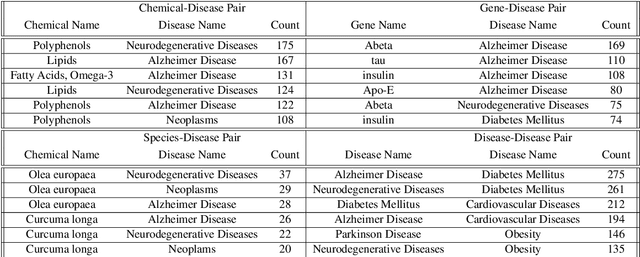

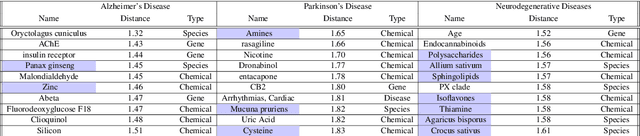
Abstract:To date, there are no effective treatments for most neurodegenerative diseases. However, certain foods may be associated with these diseases and bring an opportunity to prevent or delay neurodegenerative progression. Our objective is to construct a knowledge graph for neurodegenerative diseases using literature mining to study their relations with diet. We collected biomedical annotations (Disease, Chemical, Gene, Species, SNP&Mutation) in the abstracts from 4,300 publications relevant to both neurodegenerative diseases and diet using PubTator, an NIH-supported tool that can extract biomedical concepts from literature. A knowledge graph was created from these annotations. Graph embeddings were then trained with the node2vec algorithm to support potential concept clustering and similar concept identification. We found several food-related species and chemicals that might come from diet and have an impact on neurodegenerative diseases.
Deep Representation Learning of Patient Data from Electronic Health Records (EHR): A Systematic Review
Oct 06, 2020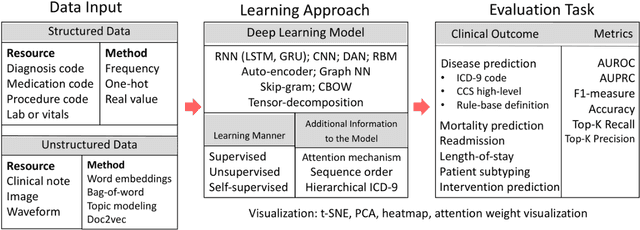
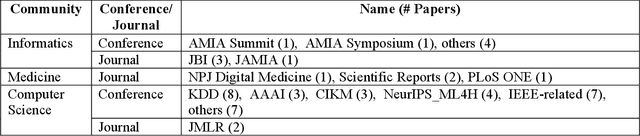
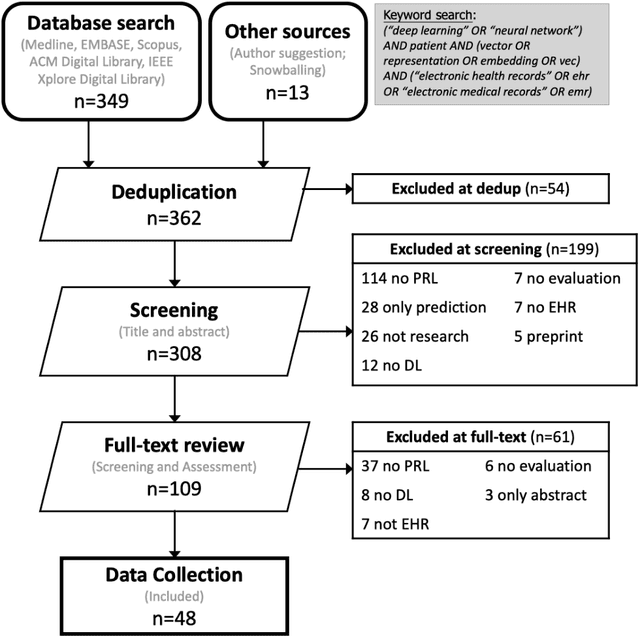
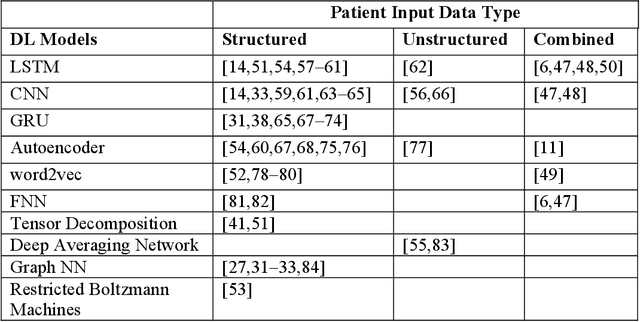
Abstract:Patient representation learning refers to learning a dense mathematical representation of a patient that encodes meaningful information from Electronic Health Records (EHRs). This is generally performed using advanced deep learning methods. This study presents a systematic review of this field and provides both qualitative and quantitative analyses from a methodological perspective. We identified studies developing patient representations from EHRs with deep learning methods from MEDLINE, EMBASE, Scopus, the Association for Computing Machinery (ACM) Digital Library, and Institute of Electrical and Electronics Engineers (IEEE) Xplore Digital Library. After screening 362 articles, 48 papers were included for a comprehensive data collection. We noticed a typical workflow starting with feeding raw data, applying deep learning models, and ending with clinical outcome predictions as evaluations of the learned representations. Specifically, learning representations from structured EHR data was dominant (36 out of 48 studies). Recurrent Neural Networks were widely applied as the deep learning architecture (LSTM: 13 studies, GRU: 11 studies). Disease prediction was the most common application and evaluation (30 studies). Benchmark datasets were mostly unavailable (28 studies) due to privacy concerns of EHR data, and code availability was assured in 20 studies. We show the importance and feasibility of learning comprehensive representations of patient EHR data through a systematic review. Advances in patient representation learning techniques will be essential for powering patient-level EHR analyses. Future work will still be devoted to leveraging the richness and potential of available EHR data. Knowledge distillation and advanced learning techniques will be exploited to assist the capability of learning patient representation further.
Deep learning with sentence embeddings pre-trained on biomedical corpora improves the performance of finding similar sentences in electronic medical records
Sep 06, 2019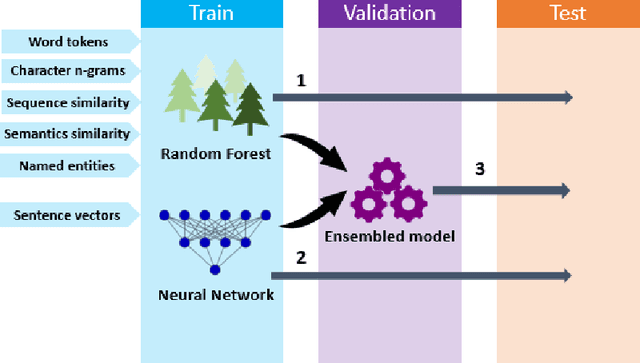
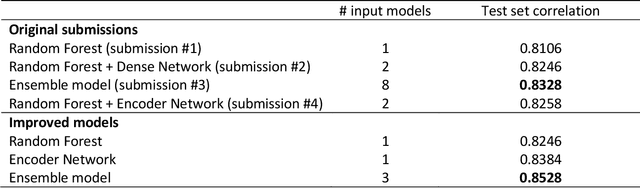
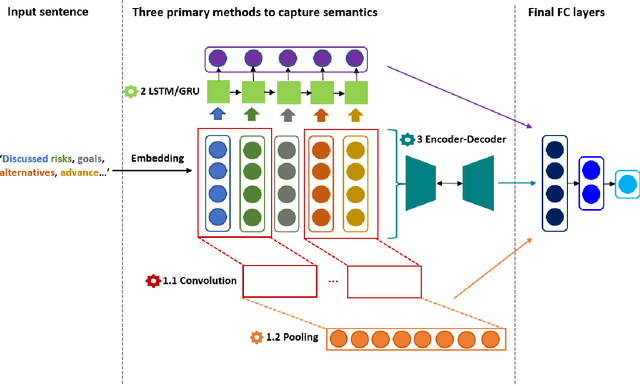
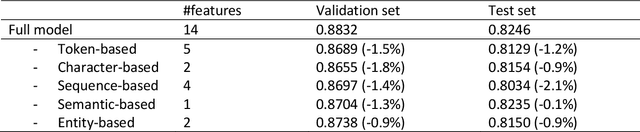
Abstract:Capturing sentence semantics plays a vital role in a range of text mining applications. Despite continuous efforts on the development of related datasets and models in the general domain, both datasets and models are limited in biomedical and clinical domains. The BioCreative/OHNLP organizers have made the first attempt to annotate 1,068 sentence pairs from clinical notes and have called for a community effort to tackle the Semantic Textual Similarity (BioCreative/OHNLP STS) challenge. We developed models using traditional machine learning and deep learning approaches. For the post challenge, we focus on two models: the Random Forest and the Encoder Network. We applied sentence embeddings pre-trained on PubMed abstracts and MIMIC-III clinical notes and updated the Random Forest and the Encoder Network accordingly. The official results demonstrated our best submission was the ensemble of eight models. It achieved a Person correlation coefficient of 0.8328, the highest performance among 13 submissions from 4 teams. For the post challenge, the performance of both Random Forest and the Encoder Network was improved; in particular, the correlation of the Encoder Network was improved by ~13%. During the challenge task, no end-to-end deep learning models had better performance than machine learning models that take manually-crafted features. In contrast, with the sentence embeddings pre-trained on biomedical corpora, the Encoder Network now achieves a correlation of ~0.84, which is higher than the original best model. The ensembled model taking the improved versions of the Random Forest and Encoder Network as inputs further increased performance to 0.8528. Deep learning models with sentence embeddings pre-trained on biomedical corpora achieve the highest performance on the test set.
Exploring difference in public perceptions on HPV vaccine between gender groups from Twitter using deep learning
Jul 06, 2019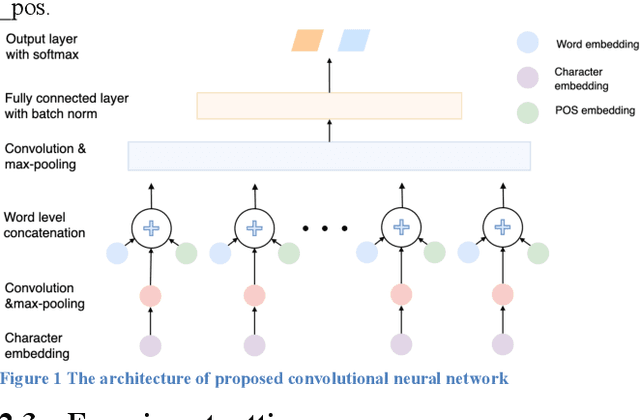
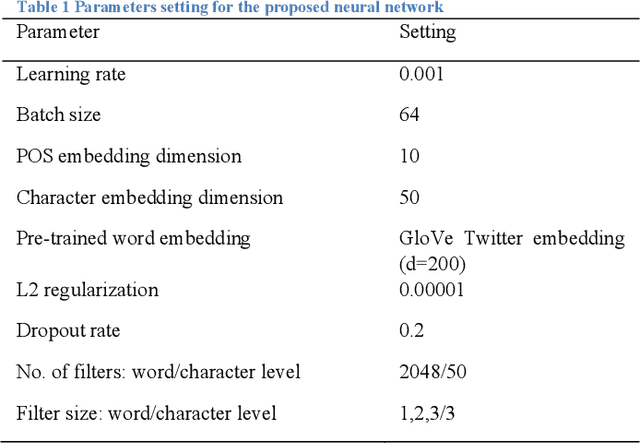
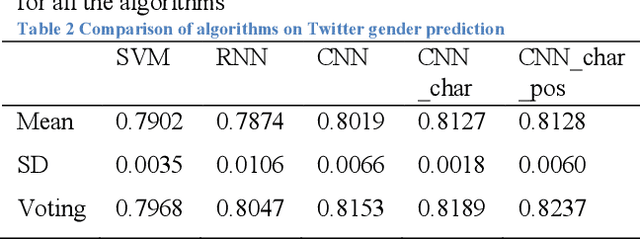
Abstract:In this study, we proposed a convolutional neural network model for gender prediction using English Twitter text as input. Ensemble of proposed model achieved an accuracy at 0.8237 on gender prediction and compared favorably with the state-of-the-art performance in a recent author profiling task. We further leveraged the trained models to predict the gender labels from an HPV vaccine related corpus and identified gender difference in public perceptions regarding HPV vaccine. The findings are largely consistent with previous survey-based studies.
ML-Net: multi-label classification of biomedical texts with deep neural networks
Nov 15, 2018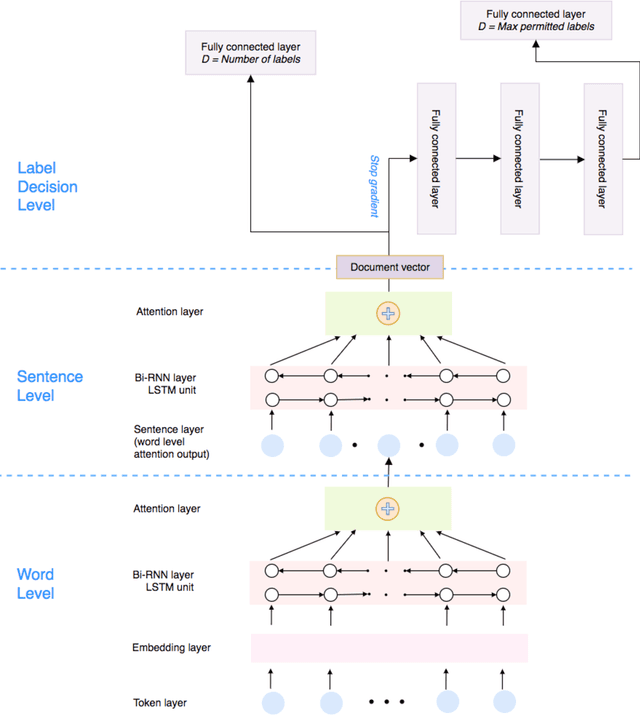
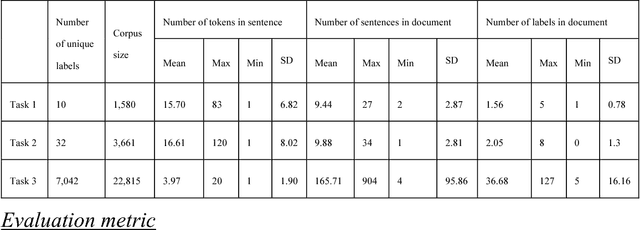
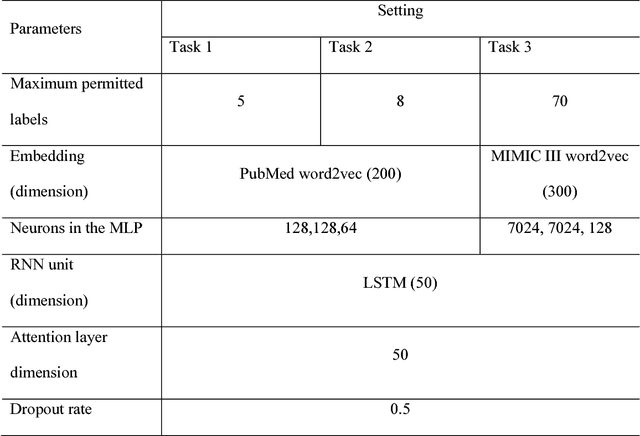
Abstract:In multi-label text classification, each textual document can be assigned with one or more labels. Due to this nature, the multi-label text classification task is often considered to be more challenging compared to the binary or multi-class text classification problems. As an important task with broad applications in biomedicine such as assigning diagnosis codes, a number of different computational methods (e.g. training and combining binary classifiers for each label) have been proposed in recent years. However, many suffered from modest accuracy and efficiency, with only limited success in practical use. We propose ML-Net, a novel deep learning framework, for multi-label classification of biomedical texts. As an end-to-end system, ML-Net combines a label prediction network with an automated label count prediction mechanism to output an optimal set of labels by leveraging both predicted confidence score of each label and the contextual information in the target document. We evaluate ML-Net on three independent, publicly-available corpora in two kinds of text genres: biomedical literature and clinical notes. For evaluation, example-based measures such as precision, recall and f-measure are used. ML-Net is compared with several competitive machine learning baseline models. Our benchmarking results show that ML-Net compares favorably to the state-of-the-art methods in multi-label classification of biomedical texts. ML-NET is also shown to be robust when evaluated on different text genres in biomedicine. Unlike traditional machine learning methods, ML-Net does not require human efforts in feature engineering and is highly efficient and scalable approach to tasks with a large set of labels (no need to build individual classifiers for each separate label). Finally, ML-NET is able to dynamically estimate the label count based on the document context in a more systematic and accurate manner.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge