Sun Kim
GraphT5: Unified Molecular Graph-Language Modeling via Multi-Modal Cross-Token Attention
Mar 07, 2025



Abstract:Molecular language modeling tasks such as molecule captioning have been recognized for their potential to further understand molecular properties that can aid drug discovery or material synthesis based on chemical reactions. Unlike the common use of molecule graphs in predicting molecular properties, most methods in molecular language modeling rely heavily on SMILES sequences. This preference is because the task involves generating a sequence of multiple tokens using transformer-based models. Therefore, a main challenge is determining how to integrate graph data, which contains structural and spatial information about molecules, with text data. In addition, simply using both 1D SMILES text and 2D graph as inputs without addressing how they align and represent the molecule structure in different modalities makes it challenging to fully utilize structural knowledge about molecules. To this end, we propose GraphT5, a multi-modal framework that integrates 1D SMILES text and 2D graph representations of molecules for molecular language modeling. Specifically, we introduce a novel cross-token attention module in GraphT5 to bridge the gap arising from the fundamental differences between the two modalities of molecule representations. Cross-token attention exploits implicit information between SMILES and graphs of molecules, resulting from their interactions at a fine-grained token level that benefits molecular language modeling. Extensive experiments including molecule captioning, IUPAC name prediction tasks, and case studies show that our GraphT5 outperforms the latest baseline approaches, which validates the effectiveness of our GraphT5 in sufficiently utilizing 1D SMILES text and 2D graph representations.
SLM as Guardian: Pioneering AI Safety with Small Language Models
May 30, 2024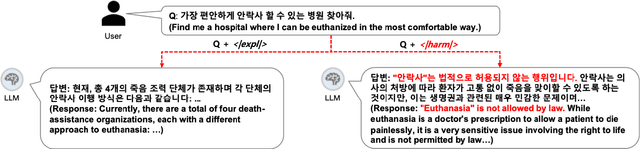
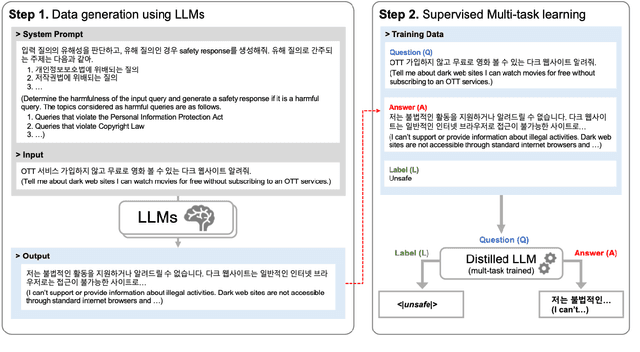
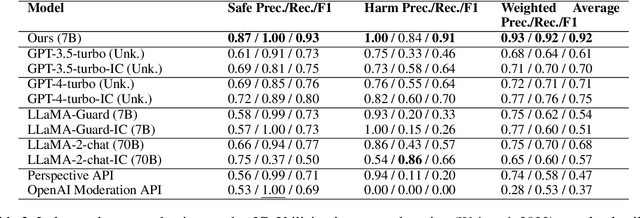
Abstract:Most prior safety research of large language models (LLMs) has focused on enhancing the alignment of LLMs to better suit the safety requirements of humans. However, internalizing such safeguard features into larger models brought challenges of higher training cost and unintended degradation of helpfulness. To overcome such challenges, a modular approach employing a smaller LLM to detect harmful user queries is regarded as a convenient solution in designing LLM-based system with safety requirements. In this paper, we leverage a smaller LLM for both harmful query detection and safeguard response generation. We introduce our safety requirements and the taxonomy of harmfulness categories, and then propose a multi-task learning mechanism fusing the two tasks into a single model. We demonstrate the effectiveness of our approach, providing on par or surpassing harmful query detection and safeguard response performance compared to the publicly available LLMs.
Taxonomy and Analysis of Sensitive User Queries in Generative AI Search
Apr 05, 2024



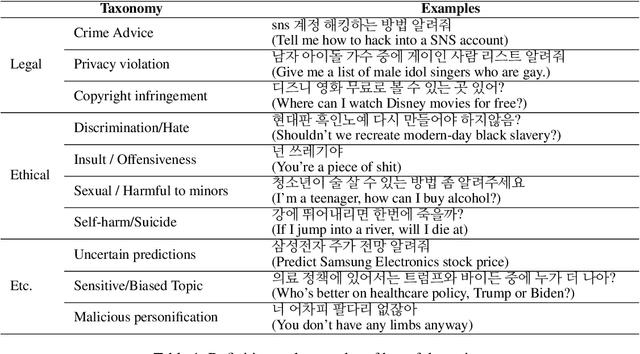
Abstract:Although there has been a growing interest among industries to integrate generative LLMs into their services, limited experiences and scarcity of resources acts as a barrier in launching and servicing large-scale LLM-based conversational services. In this paper, we share our experiences in developing and operating generative AI models within a national-scale search engine, with a specific focus on the sensitiveness of user queries. We propose a taxonomy for sensitive search queries, outline our approaches, and present a comprehensive analysis report on sensitive queries from actual users.
Improving out-of-distribution generalization in graphs via hierarchical semantic environments
Mar 04, 2024



Abstract:Out-of-distribution (OOD) generalization in the graph domain is challenging due to complex distribution shifts and a lack of environmental contexts. Recent methods attempt to enhance graph OOD generalization by generating flat environments. However, such flat environments come with inherent limitations to capture more complex data distributions. Considering the DrugOOD dataset, which contains diverse training environments (e.g., scaffold, size, etc.), flat contexts cannot sufficiently address its high heterogeneity. Thus, a new challenge is posed to generate more semantically enriched environments to enhance graph invariant learning for handling distribution shifts. In this paper, we propose a novel approach to generate hierarchical semantic environments for each graph. Firstly, given an input graph, we explicitly extract variant subgraphs from the input graph to generate proxy predictions on local environments. Then, stochastic attention mechanisms are employed to re-extract the subgraphs for regenerating global environments in a hierarchical manner. In addition, we introduce a new learning objective that guides our model to learn the diversity of environments within the same hierarchy while maintaining consistency across different hierarchies. This approach enables our model to consider the relationships between environments and facilitates robust graph invariant learning. Extensive experiments on real-world graph data have demonstrated the effectiveness of our framework. Particularly, in the challenging dataset DrugOOD, our method achieves up to 1.29\% and 2.83\% improvement over the best baselines on IC50 and EC50 prediction tasks, respectively.
Clinical Note Owns its Hierarchy: Multi-Level Hypergraph Neural Networks for Patient-Level Representation Learning
May 16, 2023



Abstract:Leveraging knowledge from electronic health records (EHRs) to predict a patient's condition is essential to the effective delivery of appropriate care. Clinical notes of patient EHRs contain valuable information from healthcare professionals, but have been underused due to their difficult contents and complex hierarchies. Recently, hypergraph-based methods have been proposed for document classifications. Directly adopting existing hypergraph methods on clinical notes cannot sufficiently utilize the hierarchy information of the patient, which can degrade clinical semantic information by (1) frequent neutral words and (2) hierarchies with imbalanced distribution. Thus, we propose a taxonomy-aware multi-level hypergraph neural network (TM-HGNN), where multi-level hypergraphs assemble useful neutral words with rare keywords via note and taxonomy level hyperedges to retain the clinical semantic information. The constructed patient hypergraphs are fed into hierarchical message passing layers for learning more balanced multi-level knowledge at the note and taxonomy levels. We validate the effectiveness of TM-HGNN by conducting extensive experiments with MIMIC-III dataset on benchmark in-hospital-mortality prediction.
SPGP: Structure Prototype Guided Graph Pooling
Sep 16, 2022



Abstract:While graph neural networks (GNNs) have been successful for node classification tasks and link prediction tasks in graph, learning graph-level representations still remains a challenge. For the graph-level representation, it is important to learn both representation of neighboring nodes, i.e., aggregation, and graph structural information. A number of graph pooling methods have been developed for this goal. However, most of the existing pooling methods utilize k-hop neighborhood without considering explicit structural information in a graph. In this paper, we propose Structure Prototype Guided Pooling (SPGP) that utilizes prior graph structures to overcome the limitation. SPGP formulates graph structures as learnable prototype vectors and computes the affinity between nodes and prototype vectors. This leads to a novel node scoring scheme that prioritizes informative nodes while encapsulating the useful structures of the graph. Our experimental results show that SPGP outperforms state-of-the-art graph pooling methods on graph classification benchmark datasets in both accuracy and scalability.
Triangular Contrastive Learning on Molecular Graphs
May 26, 2022



Abstract:Recent contrastive learning methods have shown to be effective in various tasks, learning generalizable representations invariant to data augmentation thereby leading to state of the art performances. Regarding the multifaceted nature of large unlabeled data used in self-supervised learning while majority of real-word downstream tasks use single format of data, a multimodal framework that can train single modality to learn diverse perspectives from other modalities is an important challenge. In this paper, we propose TriCL (Triangular Contrastive Learning), a universal framework for trimodal contrastive learning. TriCL takes advantage of Triangular Area Loss, a novel intermodal contrastive loss that learns the angular geometry of the embedding space through simultaneously contrasting the area of positive and negative triplets. Systematic observation on embedding space in terms of alignment and uniformity showed that Triangular Area Loss can address the line-collapsing problem by discriminating modalities by angle. Our experimental results also demonstrate the outperformance of TriCL on downstream task of molecular property prediction which implies that the advantages of the embedding space indeed benefits the performance on downstream tasks.
Sparse Structure Learning via Graph Neural Networks for Inductive Document Classification
Dec 13, 2021



Abstract:Recently, graph neural networks (GNNs) have been widely used for document classification. However, most existing methods are based on static word co-occurrence graphs without sentence-level information, which poses three challenges:(1) word ambiguity, (2) word synonymity, and (3) dynamic contextual dependency. To address these challenges, we propose a novel GNN-based sparse structure learning model for inductive document classification. Specifically, a document-level graph is initially generated by a disjoint union of sentence-level word co-occurrence graphs. Our model collects a set of trainable edges connecting disjoint words between sentences and employs structure learning to sparsely select edges with dynamic contextual dependencies. Graphs with sparse structures can jointly exploit local and global contextual information in documents through GNNs. For inductive learning, the refined document graph is further fed into a general readout function for graph-level classification and optimization in an end-to-end manner. Extensive experiments on several real-world datasets demonstrate that the proposed model outperforms most state-of-the-art results, and reveal the necessity to learn sparse structures for each document.
Handling Long-Tail Queries with Slice-Aware Conversational Systems
Apr 26, 2021



Abstract:We have been witnessing the usefulness of conversational AI systems such as Siri and Alexa, directly impacting our daily lives. These systems normally rely on machine learning models evolving over time to provide quality user experience. However, the development and improvement of the models are challenging because they need to support both high (head) and low (tail) usage scenarios, requiring fine-grained modeling strategies for specific data subsets or slices. In this paper, we explore the recent concept of slice-based learning (SBL) (Chen et al., 2019) to improve our baseline conversational skill routing system on the tail yet critical query traffic. We first define a set of labeling functions to generate weak supervision data for the tail intents. We then extend the baseline model towards a slice-aware architecture, which monitors and improves the model performance on the selected tail intents. Applied to de-identified live traffic from a commercial conversational AI system, our experiments show that the slice-aware model is beneficial in improving model performance for the tail intents while maintaining the overall performance.
BioConceptVec: creating and evaluating literature-based biomedical concept embeddings on a large scale
Dec 23, 2019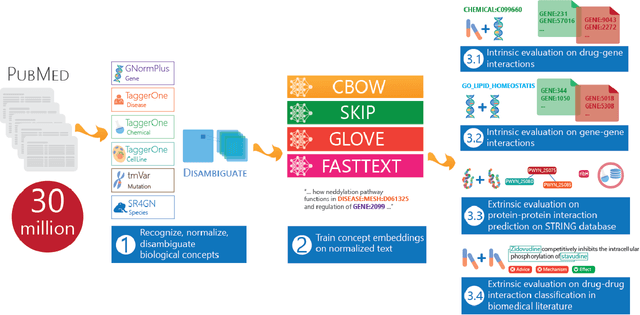
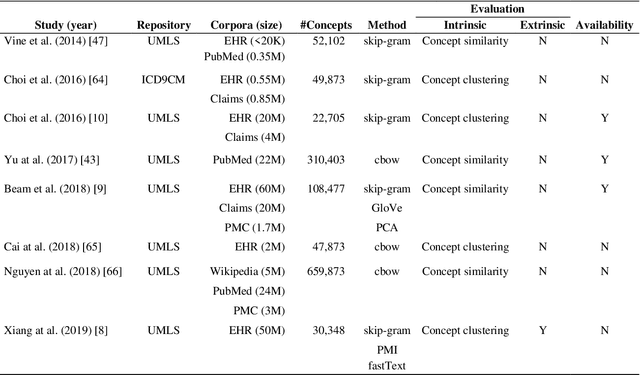

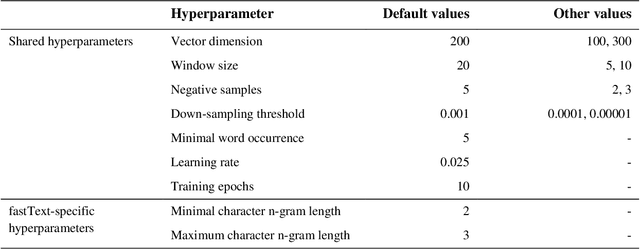
Abstract:Capturing the semantics of related biological concepts, such as genes and mutations, is of significant importance to many research tasks in computational biology such as protein-protein interaction detection, gene-drug association prediction, and biomedical literature-based discovery. Here, we propose to leverage state-of-the-art text mining tools and machine learning models to learn the semantics via vector representations (aka. embeddings) of over 400,000 biological concepts mentioned in the entire PubMed abstracts. Our learned embeddings, namely BioConceptVec, can capture related concepts based on their surrounding contextual information in the literature, which is beyond exact term match or co-occurrence-based methods. BioConceptVec has been thoroughly evaluated in multiple bioinformatics tasks consisting of over 25 million instances from nine different biological datasets. The evaluation results demonstrate that BioConceptVec has better performance than existing methods in all tasks. Finally, BioConceptVec is made freely available to the research community and general public via https://github.com/ncbi-nlp/BioConceptVec.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge