Shankai Yan
PhenoTagger: A Hybrid Method for Phenotype Concept Recognition using Human Phenotype Ontology
Sep 17, 2020



Abstract:Automatic phenotype concept recognition from unstructured text remains a challenging task in biomedical text mining research. Previous works that address the task typically use dictionary-based matching methods, which can achieve high precision but suffer from lower recall. Recently, machine learning-based methods have been proposed to identify biomedical concepts, which can recognize more unseen concept synonyms by automatic feature learning. However, most methods require large corpora of manually annotated data for model training, which is difficult to obtain due to the high cost of human annotation. In this paper, we propose PhenoTagger, a hybrid method that combines both dictionary and machine learning-based methods to recognize Human Phenotype Ontology (HPO) concepts in unstructured biomedical text. We first use all concepts and synonyms in HPO to construct a dictionary. Then, the dictionary and biomedical literature are used to automatically build a weakly-supervised training dataset for machine learning. Next, a cutting-edge deep learning model is trained to classify each candidate phrase into a corresponding concept label. Finally, the dictionary and machine learning-based prediction results are combined for improved performance. Our method is validated with two HPO corpora, and the results show that PhenoTagger compares favorably to state-of-the-art methods. In addition, to demonstrate the generalizability of our method, we retrained PhenoTagger using the disease ontology MEDIC for disease concept recognition to investigate the effect of training on different ontologies. Experimental results on the NCBI disease corpus show that PhenoTagger without requiring manually annotated training data achieves competitive performance as compared with state-of-the-art supervised methods.
BioConceptVec: creating and evaluating literature-based biomedical concept embeddings on a large scale
Dec 23, 2019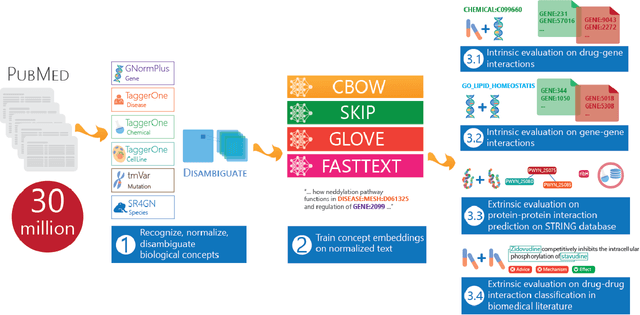
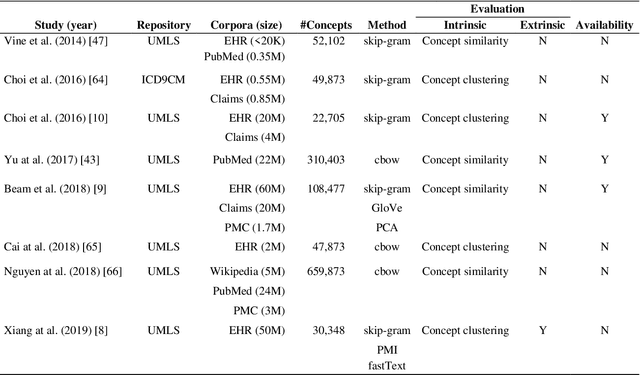

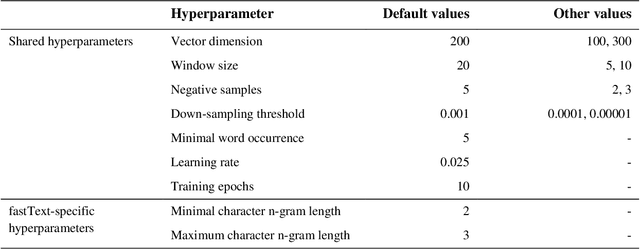
Abstract:Capturing the semantics of related biological concepts, such as genes and mutations, is of significant importance to many research tasks in computational biology such as protein-protein interaction detection, gene-drug association prediction, and biomedical literature-based discovery. Here, we propose to leverage state-of-the-art text mining tools and machine learning models to learn the semantics via vector representations (aka. embeddings) of over 400,000 biological concepts mentioned in the entire PubMed abstracts. Our learned embeddings, namely BioConceptVec, can capture related concepts based on their surrounding contextual information in the literature, which is beyond exact term match or co-occurrence-based methods. BioConceptVec has been thoroughly evaluated in multiple bioinformatics tasks consisting of over 25 million instances from nine different biological datasets. The evaluation results demonstrate that BioConceptVec has better performance than existing methods in all tasks. Finally, BioConceptVec is made freely available to the research community and general public via https://github.com/ncbi-nlp/BioConceptVec.
Transfer Learning in Biomedical Natural Language Processing: An Evaluation of BERT and ELMo on Ten Benchmarking Datasets
Jun 18, 2019


Abstract:Inspired by the success of the General Language Understanding Evaluation benchmark, we introduce the Biomedical Language Understanding Evaluation (BLUE) benchmark to facilitate research in the development of pre-training language representations in the biomedicine domain. The benchmark consists of five tasks with ten datasets that cover both biomedical and clinical texts with different dataset sizes and difficulties. We also evaluate several baselines based on BERT and ELMo and find that the BERT model pre-trained on PubMed abstracts and MIMIC-III clinical notes achieves the best results. We make the datasets, pre-trained models, and codes publicly available at https://github.com/ncbi-nlp/BLUE_Benchmark.
Context awareness and embedding for biomedical event extraction
May 02, 2019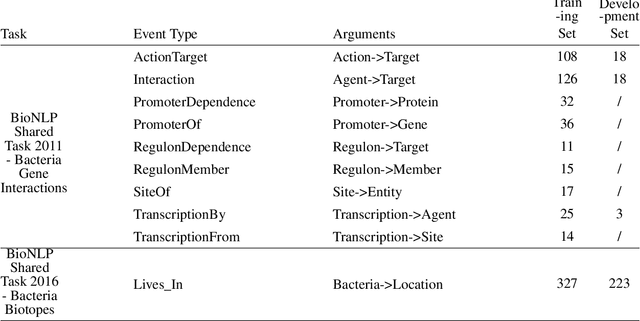
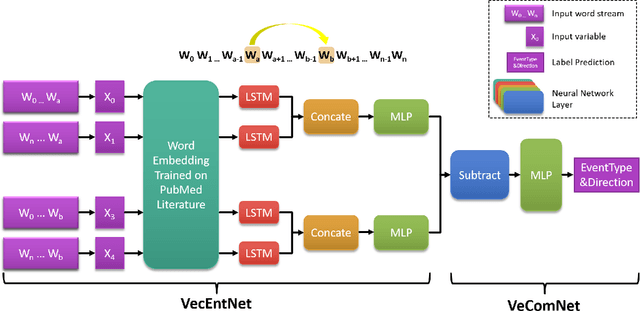
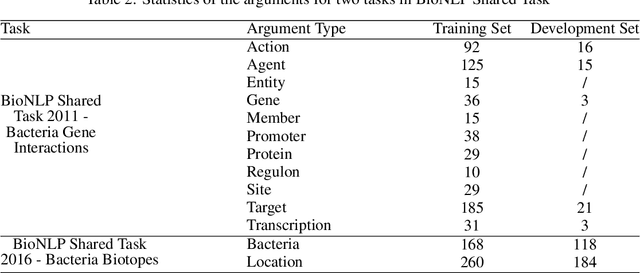
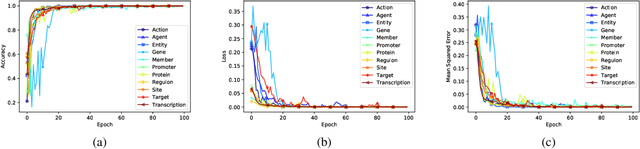
Abstract:Motivation: Biomedical event detection is fundamental for information extraction in molecular biology and biomedical research. The detected events form the central basis for comprehensive biomedical knowledge fusion, facilitating the digestion of massive information influx from literature. Limited by the feature context, the existing event detection models are mostly applicable for a single task. A general and scalable computational model is desiderated for biomedical knowledge management. Results: We consider and propose a bottom-up detection framework to identify the events from recognized arguments. To capture the relations between the arguments, we trained a bi-directional Long Short-Term Memory (LSTM) network to model their context embedding. Leveraging the compositional attributes, we further derived the candidate samples for training event classifiers. We built our models on the datasets from BioNLP Shared Task for evaluations. Our method achieved the average F-scores of 0.81 and 0.92 on BioNLPST-BGI and BioNLPST-BB datasets respectively. Comparing with 7 state-of-the-art methods, our method nearly doubled the existing F-score performance (0.92 vs 0.56) on the BioNLPST-BB dataset. Case studies were conducted to reveal the underlying reasons. Availability: https://github.com/cskyan/evntextrc
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge