Kyubum Lee
BioConceptVec: creating and evaluating literature-based biomedical concept embeddings on a large scale
Dec 23, 2019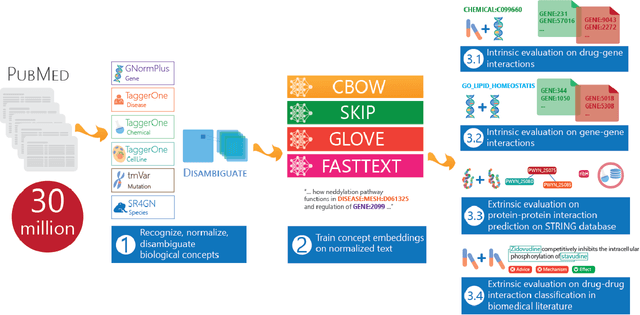
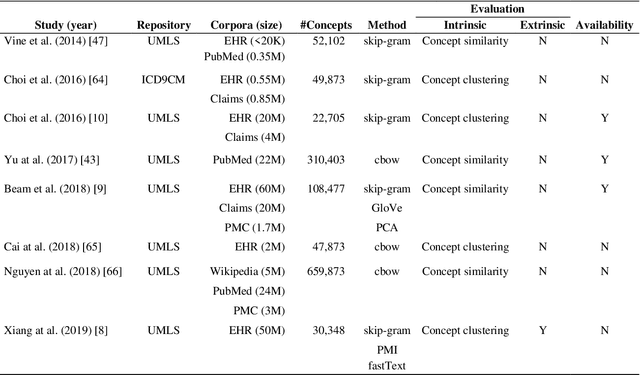

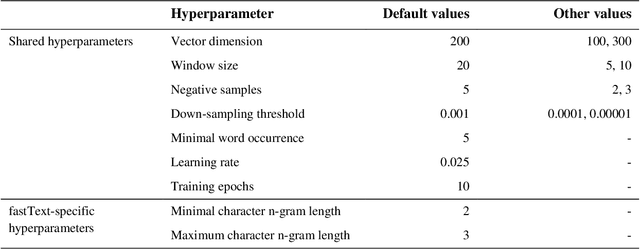
Abstract:Capturing the semantics of related biological concepts, such as genes and mutations, is of significant importance to many research tasks in computational biology such as protein-protein interaction detection, gene-drug association prediction, and biomedical literature-based discovery. Here, we propose to leverage state-of-the-art text mining tools and machine learning models to learn the semantics via vector representations (aka. embeddings) of over 400,000 biological concepts mentioned in the entire PubMed abstracts. Our learned embeddings, namely BioConceptVec, can capture related concepts based on their surrounding contextual information in the literature, which is beyond exact term match or co-occurrence-based methods. BioConceptVec has been thoroughly evaluated in multiple bioinformatics tasks consisting of over 25 million instances from nine different biological datasets. The evaluation results demonstrate that BioConceptVec has better performance than existing methods in all tasks. Finally, BioConceptVec is made freely available to the research community and general public via https://github.com/ncbi-nlp/BioConceptVec.
Biomedical Mention Disambiguation using a Deep Learning Approach
Sep 23, 2019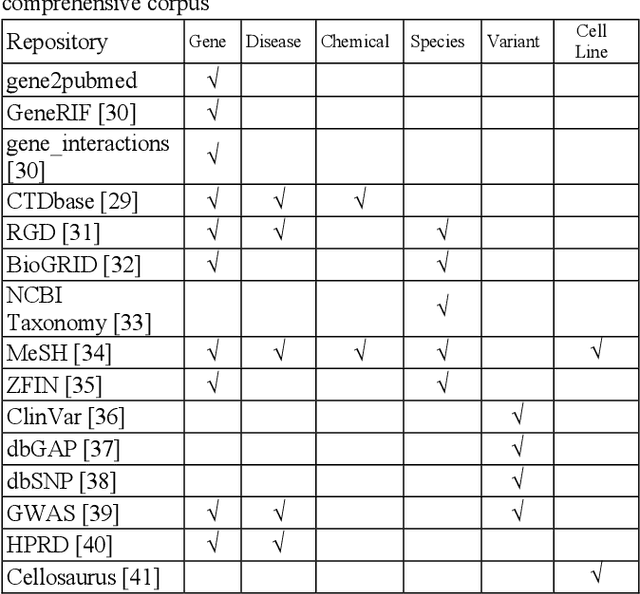
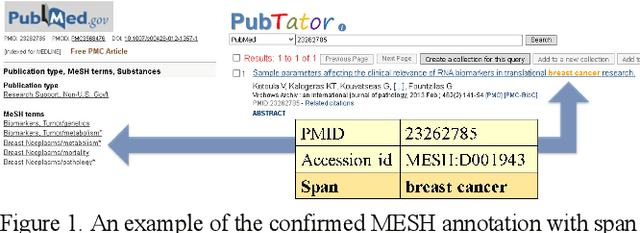
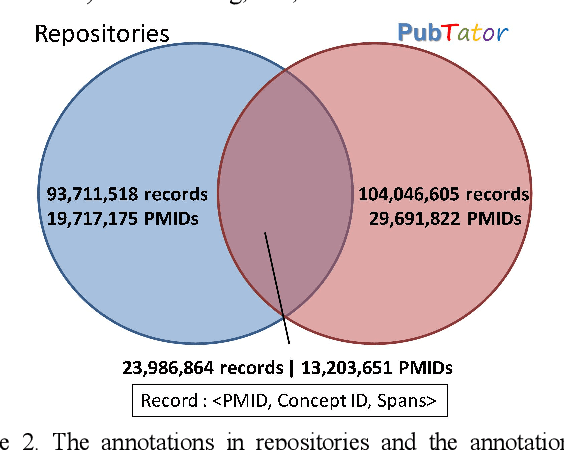
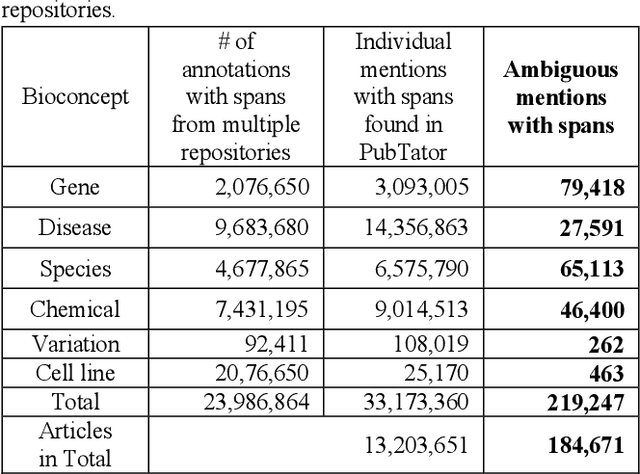
Abstract:Automatically locating named entities in natural language text - named entity recognition - is an important task in the biomedical domain. Many named entity mentions are ambiguous between several bioconcept types, however, causing text spans to be annotated as more than one type when simultaneously recognizing multiple entity types. The straightforward solution is a rule-based approach applying a priority order based on the precision of each entity tagger (from highest to lowest). While this method is straightforward and useful, imprecise disambiguation remains a significant source of error. We address this issue by generating a partially labeled corpus of ambiguous concept mentions. We first collect named entity mentions from multiple human-curated databases (e.g. CTDbase, gene2pubmed), then correlate them with the text mined span from PubTator to provide the context where the mention appears. Our corpus contains more than 3 million concept mentions that ambiguous between one or more concept types in PubTator (about 3% of all mentions). We approached this task as a classification problem and developed a deep learning-based method which uses the semantics of the span being classified and the surrounding words to identify the most likely bioconcept type. More specifically, we develop a convolutional neural network (CNN) and along short-term memory (LSTM) network to respectively handle the semantic syntax features, then concatenate these within a fully connected layer for final classification. The priority ordering rule-based approach demonstrated F1-scores of 71.29% (micro-averaged) and 41.19% (macro-averaged), while the new disambiguation method demonstrated F1-scores of 91.94% (micro-averaged) and 85.42% (macro-averaged), a very substantial increase.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge