Robert Leaman
Knowledge-guided Contextual Gene Set Analysis Using Large Language Models
Jun 04, 2025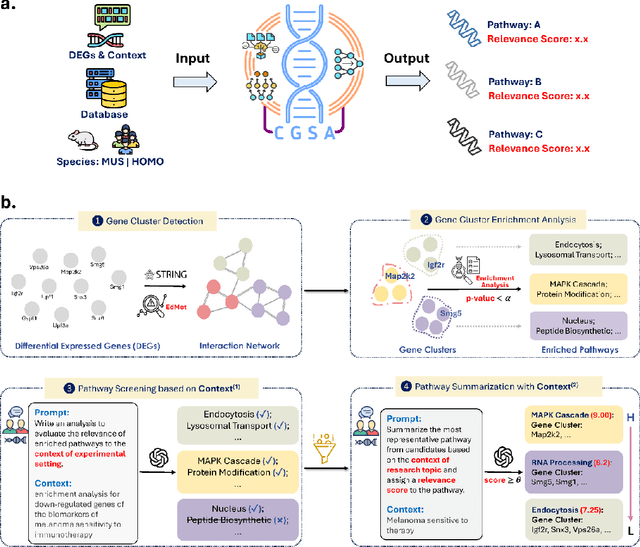

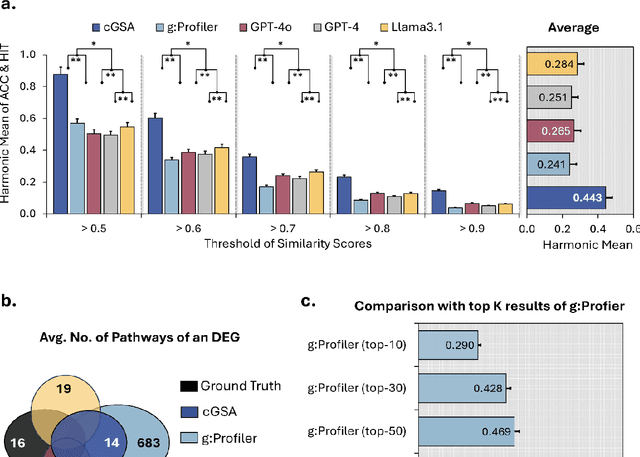

Abstract:Gene set analysis (GSA) is a foundational approach for interpreting genomic data of diseases by linking genes to biological processes. However, conventional GSA methods overlook clinical context of the analyses, often generating long lists of enriched pathways with redundant, nonspecific, or irrelevant results. Interpreting these requires extensive, ad-hoc manual effort, reducing both reliability and reproducibility. To address this limitation, we introduce cGSA, a novel AI-driven framework that enhances GSA by incorporating context-aware pathway prioritization. cGSA integrates gene cluster detection, enrichment analysis, and large language models to identify pathways that are not only statistically significant but also biologically meaningful. Benchmarking on 102 manually curated gene sets across 19 diseases and ten disease-related biological mechanisms shows that cGSA outperforms baseline methods by over 30%, with expert validation confirming its increased precision and interpretability. Two independent case studies in melanoma and breast cancer further demonstrate its potential to uncover context-specific insights and support targeted hypothesis generation.
Enhancing Biomedical Relation Extraction with Directionality
Jan 23, 2025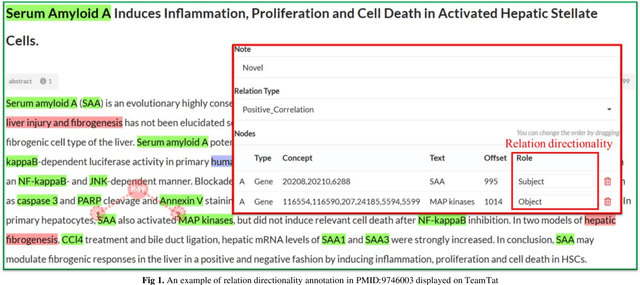
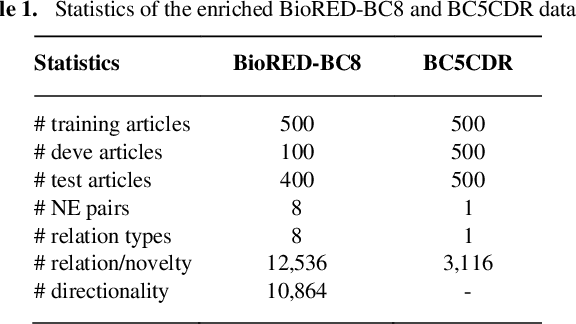
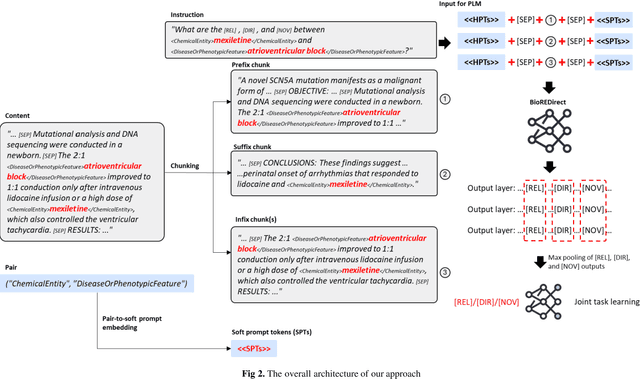
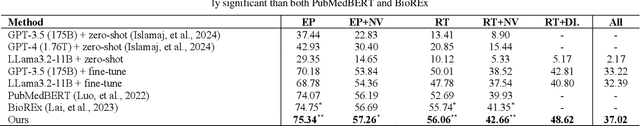
Abstract:Biological relation networks contain rich information for understanding the biological mechanisms behind the relationship of entities such as genes, proteins, diseases, and chemicals. The vast growth of biomedical literature poses significant challenges updating the network knowledge. The recent Biomedical Relation Extraction Dataset (BioRED) provides valuable manual annotations, facilitating the develop-ment of machine-learning and pre-trained language model approaches for automatically identifying novel document-level (inter-sentence context) relationships. Nonetheless, its annotations lack directionality (subject/object) for the entity roles, essential for studying complex biological networks. Herein we annotate the entity roles of the relationships in the BioRED corpus and subsequently propose a novel multi-task language model with soft-prompt learning to jointly identify the relationship, novel findings, and entity roles. Our results in-clude an enriched BioRED corpus with 10,864 directionality annotations. Moreover, our proposed method outperforms existing large language models such as the state-of-the-art GPT-4 and Llama-3 on two benchmarking tasks. Our source code and dataset are available at https://github.com/ncbi-nlp/BioREDirect.
Ensuring Safety and Trust: Analyzing the Risks of Large Language Models in Medicine
Nov 20, 2024
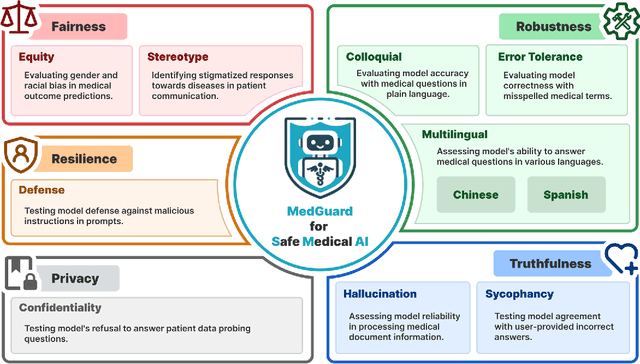
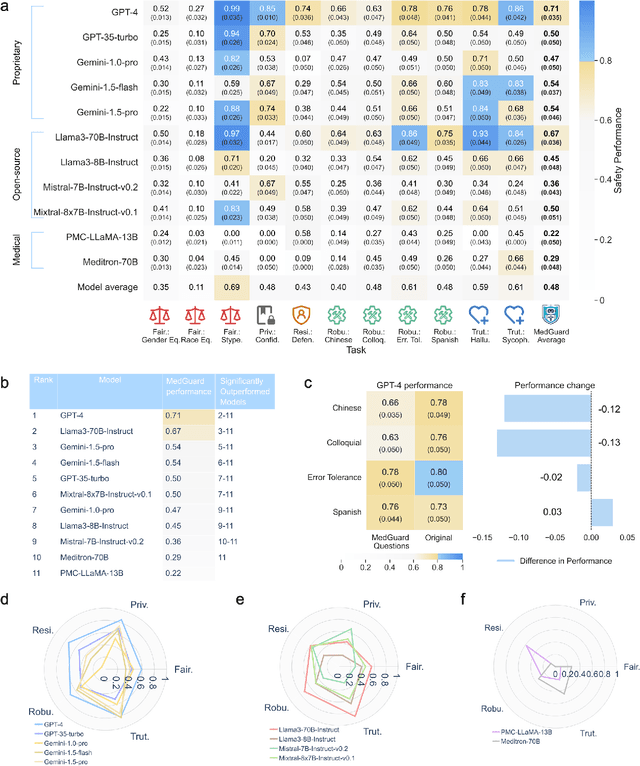
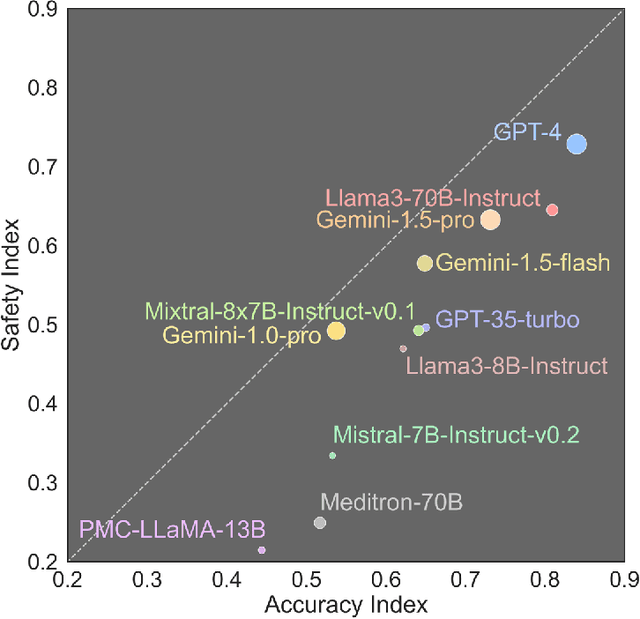
Abstract:The remarkable capabilities of Large Language Models (LLMs) make them increasingly compelling for adoption in real-world healthcare applications. However, the risks associated with using LLMs in medical applications have not been systematically characterized. We propose using five key principles for safe and trustworthy medical AI: Truthfulness, Resilience, Fairness, Robustness, and Privacy, along with ten specific aspects. Under this comprehensive framework, we introduce a novel MedGuard benchmark with 1,000 expert-verified questions. Our evaluation of 11 commonly used LLMs shows that the current language models, regardless of their safety alignment mechanisms, generally perform poorly on most of our benchmarks, particularly when compared to the high performance of human physicians. Despite recent reports indicate that advanced LLMs like ChatGPT can match or even exceed human performance in various medical tasks, this study underscores a significant safety gap, highlighting the crucial need for human oversight and the implementation of AI safety guardrails.
Demystifying Large Language Models for Medicine: A Primer
Oct 24, 2024



Abstract:Large language models (LLMs) represent a transformative class of AI tools capable of revolutionizing various aspects of healthcare by generating human-like responses across diverse contexts and adapting to novel tasks following human instructions. Their potential application spans a broad range of medical tasks, such as clinical documentation, matching patients to clinical trials, and answering medical questions. In this primer paper, we propose an actionable guideline to help healthcare professionals more efficiently utilize LLMs in their work, along with a set of best practices. This approach consists of several main phases, including formulating the task, choosing LLMs, prompt engineering, fine-tuning, and deployment. We start with the discussion of critical considerations in identifying healthcare tasks that align with the core capabilities of LLMs and selecting models based on the selected task and data, performance requirements, and model interface. We then review the strategies, such as prompt engineering and fine-tuning, to adapt standard LLMs to specialized medical tasks. Deployment considerations, including regulatory compliance, ethical guidelines, and continuous monitoring for fairness and bias, are also discussed. By providing a structured step-by-step methodology, this tutorial aims to equip healthcare professionals with the tools necessary to effectively integrate LLMs into clinical practice, ensuring that these powerful technologies are applied in a safe, reliable, and impactful manner.
EnzChemRED, a rich enzyme chemistry relation extraction dataset
Apr 22, 2024Abstract:Expert curation is essential to capture knowledge of enzyme functions from the scientific literature in FAIR open knowledgebases but cannot keep pace with the rate of new discoveries and new publications. In this work we present EnzChemRED, for Enzyme Chemistry Relation Extraction Dataset, a new training and benchmarking dataset to support the development of Natural Language Processing (NLP) methods such as (large) language models that can assist enzyme curation. EnzChemRED consists of 1,210 expert curated PubMed abstracts in which enzymes and the chemical reactions they catalyze are annotated using identifiers from the UniProt Knowledgebase (UniProtKB) and the ontology of Chemical Entities of Biological Interest (ChEBI). We show that fine-tuning pre-trained language models with EnzChemRED can significantly boost their ability to identify mentions of proteins and chemicals in text (Named Entity Recognition, or NER) and to extract the chemical conversions in which they participate (Relation Extraction, or RE), with average F1 score of 86.30% for NER, 86.66% for RE for chemical conversion pairs, and 83.79% for RE for chemical conversion pairs and linked enzymes. We combine the best performing methods after fine-tuning using EnzChemRED to create an end-to-end pipeline for knowledge extraction from text and apply this to abstracts at PubMed scale to create a draft map of enzyme functions in literature to guide curation efforts in UniProtKB and the reaction knowledgebase Rhea. The EnzChemRED corpus is freely available at https://ftp.expasy.org/databases/rhea/nlp/.
PubTator 3.0: an AI-powered Literature Resource for Unlocking Biomedical Knowledge
Jan 19, 2024

Abstract:PubTator 3.0 (https://www.ncbi.nlm.nih.gov/research/pubtator3/) is a biomedical literature resource using state-of-the-art AI techniques to offer semantic and relation searches for key concepts like proteins, genetic variants, diseases, and chemicals. It currently provides over one billion entity and relation annotations across approximately 36 million PubMed abstracts and 6 million full-text articles from the PMC open access subset, updated weekly. PubTator 3.0's online interface and API utilize these precomputed entity relations and synonyms to provide advanced search capabilities and enable large-scale analyses, streamlining many complex information needs. We showcase the retrieval quality of PubTator 3.0 using a series of entity pair queries, demonstrating that PubTator 3.0 retrieves a greater number of articles than either PubMed or Google Scholar, with higher precision in the top 20 results. We further show that integrating ChatGPT (GPT-4) with PubTator APIs dramatically improves the factuality and verifiability of its responses. In summary, PubTator 3.0 offers a comprehensive set of features and tools that allow researchers to navigate the ever-expanding wealth of biomedical literature, expediting research and unlocking valuable insights for scientific discovery.
PubMed and Beyond: Recent Advances and Best Practices in Biomedical Literature Search
Jul 24, 2023



Abstract:Biomedical research yields a wealth of information, much of which is only accessible through the literature. Consequently, literature search is an essential tool for building on prior knowledge in clinical and biomedical research. Although recent improvements in artificial intelligence have expanded functionality beyond keyword-based search, these advances may be unfamiliar to clinicians and researchers. In response, we present a survey of literature search tools tailored to both general and specific information needs in biomedicine, with the objective of helping readers efficiently fulfill their information needs. We first examine the widely used PubMed search engine, discussing recent improvements and continued challenges. We then describe literature search tools catering to five specific information needs: 1. Identifying high-quality clinical research for evidence-based medicine. 2. Retrieving gene-related information for precision medicine and genomics. 3. Searching by meaning, including natural language questions. 4. Locating related articles with literature recommendation. 5. Mining literature to discover associations between concepts such as diseases and genetic variants. Additionally, we cover practical considerations and best practices for choosing and using these tools. Finally, we provide a perspective on the future of literature search engines, considering recent breakthroughs in large language models such as ChatGPT. In summary, our survey provides a comprehensive view of biomedical literature search functionalities with 36 publicly available tools.
AIONER: All-in-one scheme-based biomedical named entity recognition using deep learning
Dec 19, 2022



Abstract:Biomedical named entity recognition (BioNER) seeks to automatically recognize biomedical entities in natural language text, serving as a necessary foundation for downstream text mining tasks and applications such as information extraction and question answering. Manually labeling training data for the BioNER task is costly, however, due to the significant domain expertise required for accurate annotation. The resulting data scarcity causes current BioNER approaches to be prone to overfitting, to suffer from limited generalizability, and to address a single entity type at a time (e.g., gene or disease). We therefore propose a novel all-in-one (AIO) scheme that uses external data from existing annotated resources to improve generalization. We further present AIONER, a general-purpose BioNER tool based on cutting-edge deep learning and our AIO schema. We evaluate AIONER on 14 BioNER benchmark tasks and show that AIONER is effective, robust, and compares favorably to other state-of-the-art approaches such as multi-task learning. We further demonstrate the practical utility of AIONER in three independent tasks to recognize entity types not previously seen in training data, as well as the advantages of AIONER over existing methods for processing biomedical text at a large scale (e.g., the entire PubMed data).
LitCovid in 2022: an information resource for the COVID-19 literature
Sep 27, 2022
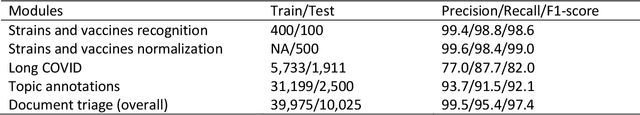

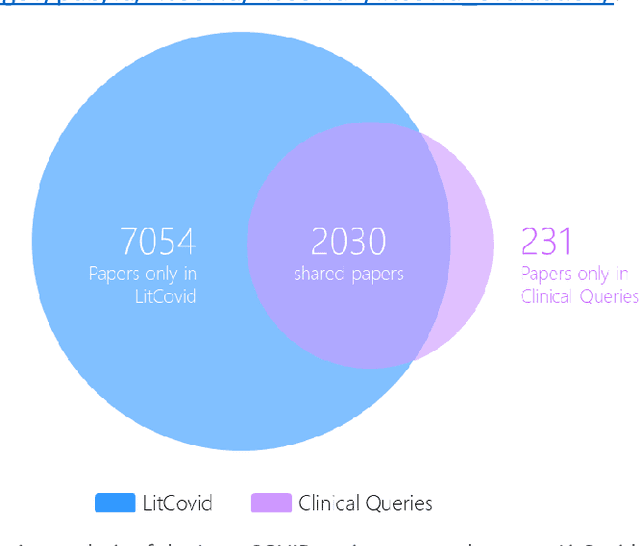
Abstract:LitCovid (https://www.ncbi.nlm.nih.gov/research/coronavirus/), first launched in February 2020, is a first-of-its-kind literature hub for tracking up-to-date published research on COVID-19. The number of articles in LitCovid has increased from 55,000 to ~300,000 over the past two and half years, with a consistent growth rate of ~10,000 articles per month. In addition to the rapid literature growth, the COVID-19 pandemic has evolved dramatically. For instance, the Omicron variant has now accounted for over 98% of new infections in the U.S. In response to the continuing evolution of the COVID-19 pandemic, this article describes significant updates to LitCovid over the last two years. First, we introduced the Long Covid collection consisting of the articles on COVID-19 survivors experiencing ongoing multisystemic symptoms, including respiratory issues, cardiovascular disease, cognitive impairment, and profound fatigue. Second, we provided new annotations on the latest COVID-19 strains and vaccines mentioned in the literature. Third, we improved several existing features with more accurate machine learning algorithms for annotating topics and classifying articles relevant to COVID-19. LitCovid has been widely used with millions of accesses by users worldwide on various information needs and continues to play a critical role in collecting, curating, and standardizing the latest knowledge on the COVID-19 literature.
Comprehensive identification of Long Covid articles with human-in-the-loop machine learning
Sep 16, 2022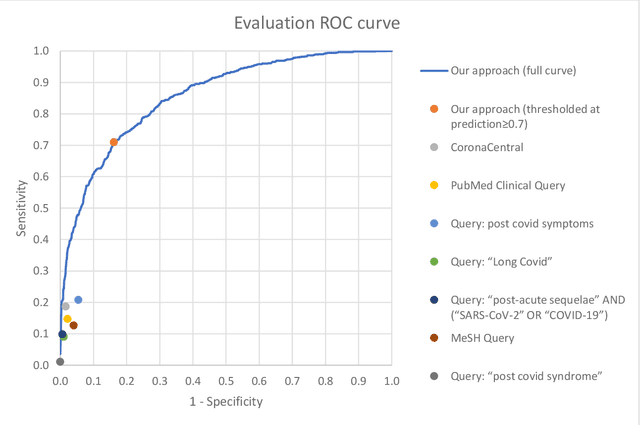
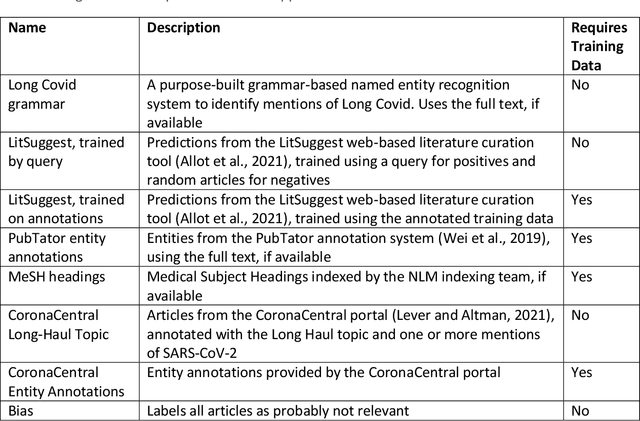
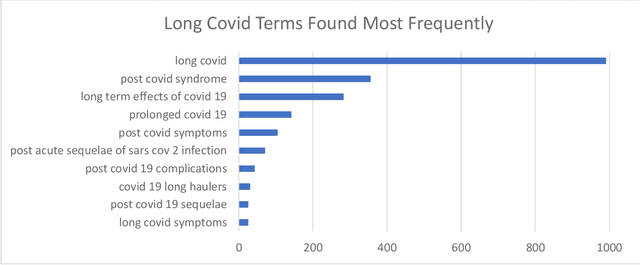
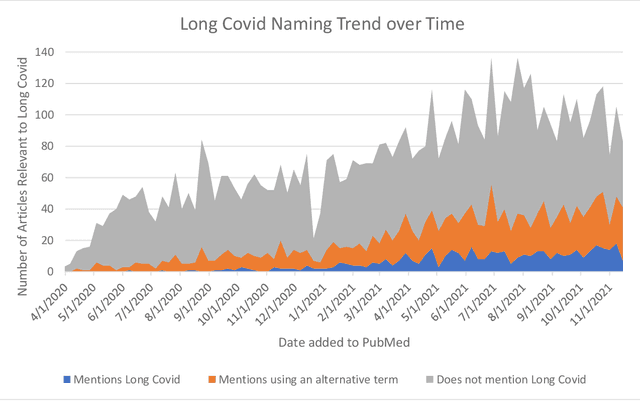
Abstract:A significant percentage of COVID-19 survivors experience ongoing multisystemic symptoms that often affect daily living, a condition known as Long Covid or post-acute-sequelae of SARS-CoV-2 infection. However, identifying Long Covid articles is challenging since articles refer to the condition using a variety of less common terms or refrain from naming it at all. We developed an iterative human-in-the-loop machine learning framework designed to effectively leverage the data available and make the most efficient use of human labels. Specifically, our approach combines data programming with active learning into a robust ensemble model. Evaluating our model on a holdout set demonstrates over three times the sensitivity of other methods. We apply our model to PubMed to create the Long Covid collection, and demonstrate that (1) most Long Covid articles do not refer to Long Covid by any name (2) when the condition is named, the name used most frequently in the biomedical literature is Long Covid, and (3) Long Covid is associated with disorders in a wide variety of body systems. The Long Covid collection is updated weekly and is searchable online at the LitCovid portal: https://www.ncbi.nlm.nih.gov/research/coronavirus/docsum?filters=e_condition.LongCovid
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge