Yin Fang
Knowledge-guided Contextual Gene Set Analysis Using Large Language Models
Jun 04, 2025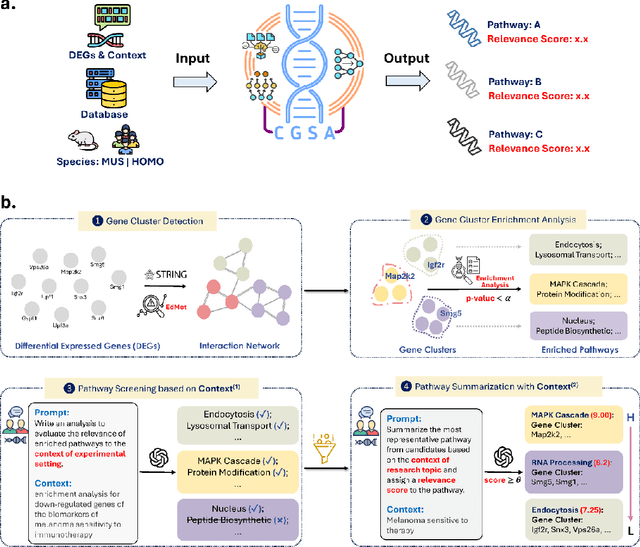

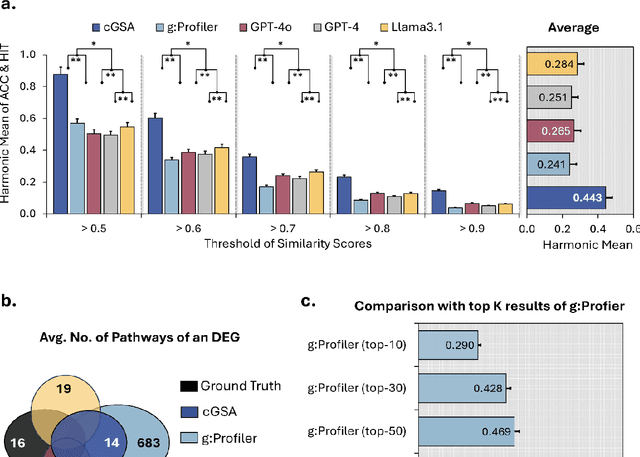

Abstract:Gene set analysis (GSA) is a foundational approach for interpreting genomic data of diseases by linking genes to biological processes. However, conventional GSA methods overlook clinical context of the analyses, often generating long lists of enriched pathways with redundant, nonspecific, or irrelevant results. Interpreting these requires extensive, ad-hoc manual effort, reducing both reliability and reproducibility. To address this limitation, we introduce cGSA, a novel AI-driven framework that enhances GSA by incorporating context-aware pathway prioritization. cGSA integrates gene cluster detection, enrichment analysis, and large language models to identify pathways that are not only statistically significant but also biologically meaningful. Benchmarking on 102 manually curated gene sets across 19 diseases and ten disease-related biological mechanisms shows that cGSA outperforms baseline methods by over 30%, with expert validation confirming its increased precision and interpretability. Two independent case studies in melanoma and breast cancer further demonstrate its potential to uncover context-specific insights and support targeted hypothesis generation.
Multi-Scale Target-Aware Representation Learning for Fundus Image Enhancement
May 03, 2025Abstract:High-quality fundus images provide essential anatomical information for clinical screening and ophthalmic disease diagnosis. Yet, due to hardware limitations, operational variability, and patient compliance, fundus images often suffer from low resolution and signal-to-noise ratio. Recent years have witnessed promising progress in fundus image enhancement. However, existing works usually focus on restoring structural details or global characteristics of fundus images, lacking a unified image enhancement framework to recover comprehensive multi-scale information. Moreover, few methods pinpoint the target of image enhancement, e.g., lesions, which is crucial for medical image-based diagnosis. To address these challenges, we propose a multi-scale target-aware representation learning framework (MTRL-FIE) for efficient fundus image enhancement. Specifically, we propose a multi-scale feature encoder (MFE) that employs wavelet decomposition to embed both low-frequency structural information and high-frequency details. Next, we design a structure-preserving hierarchical decoder (SHD) to fuse multi-scale feature embeddings for real fundus image restoration. SHD integrates hierarchical fusion and group attention mechanisms to achieve adaptive feature fusion while retaining local structural smoothness. Meanwhile, a target-aware feature aggregation (TFA) module is used to enhance pathological regions and reduce artifacts. Experimental results on multiple fundus image datasets demonstrate the effectiveness and generalizability of MTRL-FIE for fundus image enhancement. Compared to state-of-the-art methods, MTRL-FIE achieves superior enhancement performance with a more lightweight architecture. Furthermore, our approach generalizes to other ophthalmic image processing tasks without supervised fine-tuning, highlighting its potential for clinical applications.
RAG-Gym: Optimizing Reasoning and Search Agents with Process Supervision
Feb 19, 2025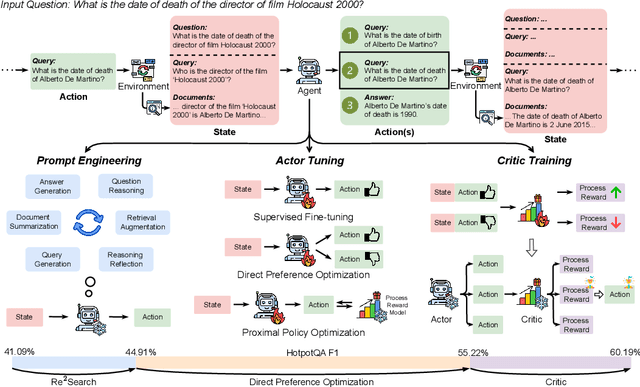

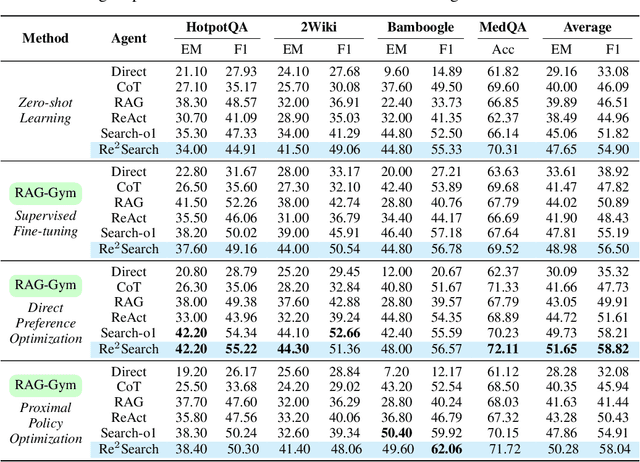
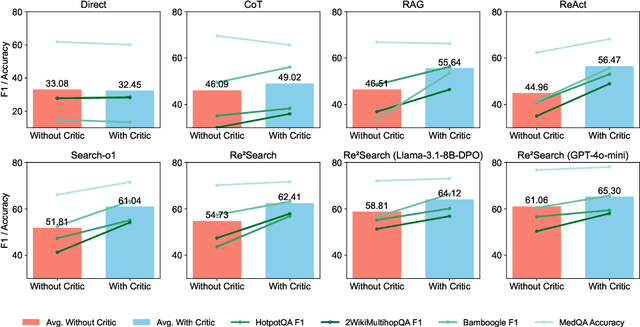
Abstract:Retrieval-augmented generation (RAG) has shown great potential for knowledge-intensive tasks, but its traditional architectures rely on static retrieval, limiting their effectiveness for complex questions that require sequential information-seeking. While agentic reasoning and search offer a more adaptive approach, most existing methods depend heavily on prompt engineering. In this work, we introduce RAG-Gym, a unified optimization framework that enhances information-seeking agents through fine-grained process supervision at each search step. We also propose ReSearch, a novel agent architecture that synergizes answer reasoning and search query generation within the RAG-Gym framework. Experiments on four challenging datasets show that RAG-Gym improves performance by up to 25.6\% across various agent architectures, with ReSearch consistently outperforming existing baselines. Further analysis highlights the effectiveness of advanced LLMs as process reward judges and the transferability of trained reward models as verifiers for different LLMs. Additionally, we examine the scaling properties of training and inference in agentic RAG. The project homepage is available at https://rag-gym.github.io/.
A Multi-Modal AI Copilot for Single-Cell Analysis with Instruction Following
Jan 15, 2025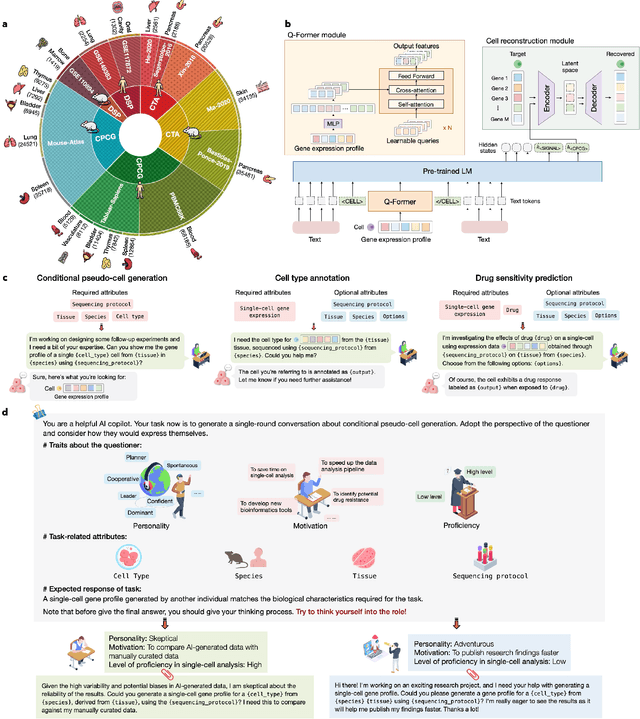
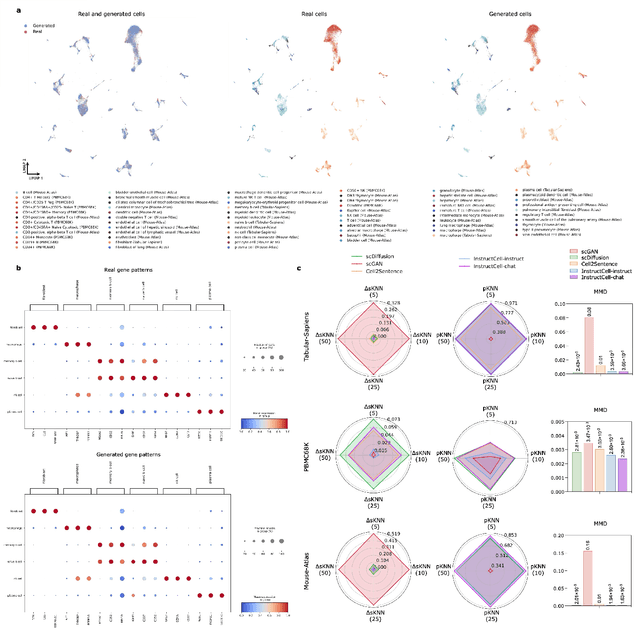
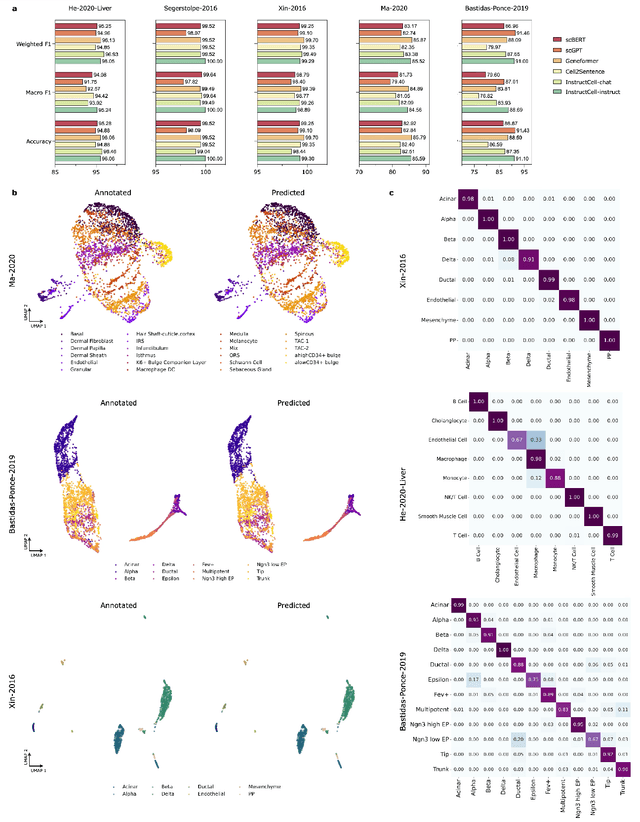
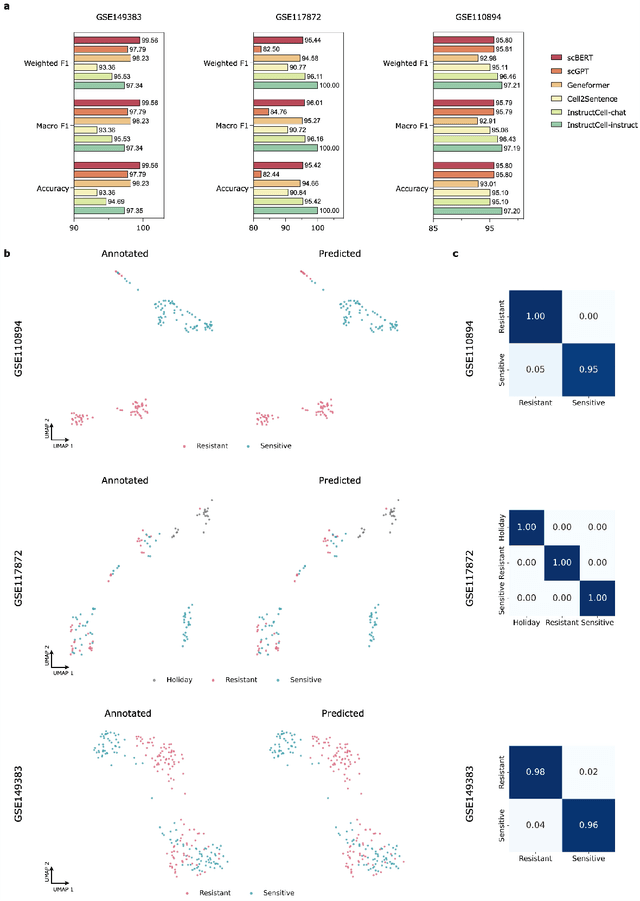
Abstract:Large language models excel at interpreting complex natural language instructions, enabling them to perform a wide range of tasks. In the life sciences, single-cell RNA sequencing (scRNA-seq) data serves as the "language of cellular biology", capturing intricate gene expression patterns at the single-cell level. However, interacting with this "language" through conventional tools is often inefficient and unintuitive, posing challenges for researchers. To address these limitations, we present InstructCell, a multi-modal AI copilot that leverages natural language as a medium for more direct and flexible single-cell analysis. We construct a comprehensive multi-modal instruction dataset that pairs text-based instructions with scRNA-seq profiles from diverse tissues and species. Building on this, we develop a multi-modal cell language architecture capable of simultaneously interpreting and processing both modalities. InstructCell empowers researchers to accomplish critical tasks-such as cell type annotation, conditional pseudo-cell generation, and drug sensitivity prediction-using straightforward natural language commands. Extensive evaluations demonstrate that InstructCell consistently meets or exceeds the performance of existing single-cell foundation models, while adapting to diverse experimental conditions. More importantly, InstructCell provides an accessible and intuitive tool for exploring complex single-cell data, lowering technical barriers and enabling deeper biological insights.
The Power of Noise: Toward a Unified Multi-modal Knowledge Graph Representation Framework
Mar 20, 2024
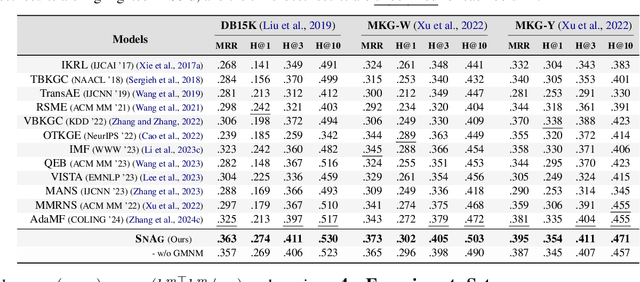
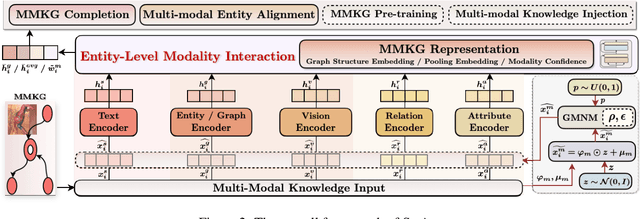
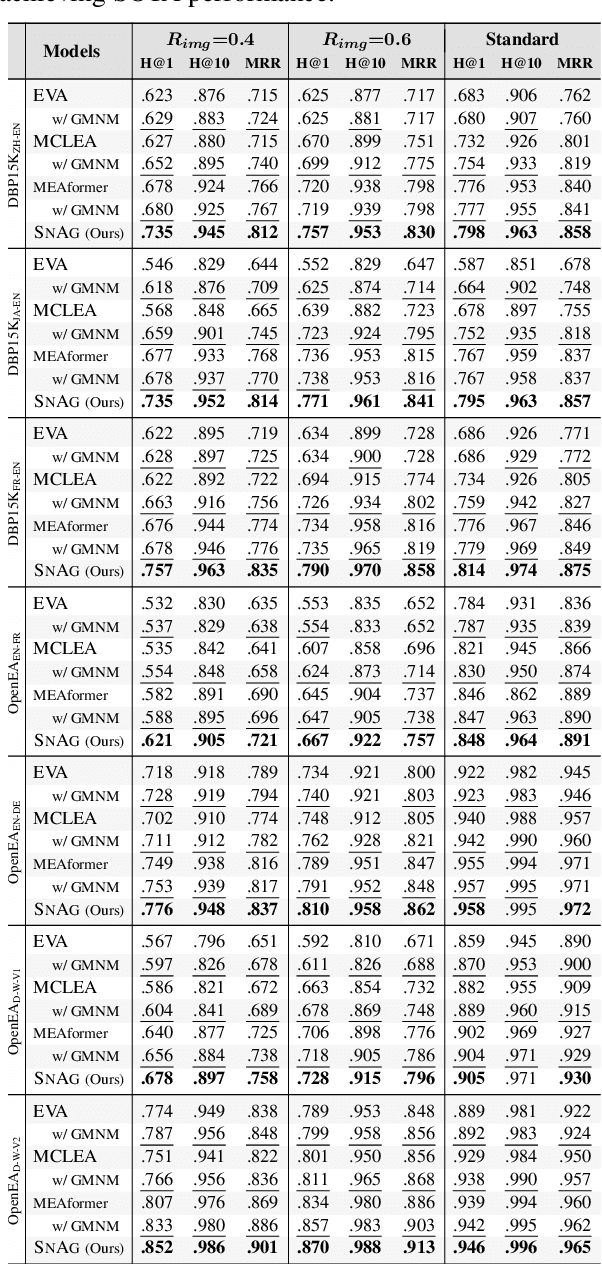
Abstract:The advancement of Multi-modal Pre-training highlights the necessity for a robust Multi-Modal Knowledge Graph (MMKG) representation learning framework. This framework is crucial for integrating structured knowledge into multi-modal Large Language Models (LLMs) at scale, aiming to alleviate issues like knowledge misconceptions and multi-modal hallucinations. In this work, to evaluate models' ability to accurately embed entities within MMKGs, we focus on two widely researched tasks: Multi-modal Knowledge Graph Completion (MKGC) and Multi-modal Entity Alignment (MMEA). Building on this foundation, we propose a novel SNAG method that utilizes a Transformer-based architecture equipped with modality-level noise masking for the robust integration of multi-modal entity features in KGs. By incorporating specific training objectives for both MKGC and MMEA, our approach achieves SOTA performance across a total of ten datasets (three for MKGC and seven for MEMA), demonstrating its robustness and versatility. Besides, SNAG can not only function as a standalone model but also enhance other existing methods, providing stable performance improvements. Our code and data are available at: https://github.com/zjukg/SNAG.
BioT5+: Towards Generalized Biological Understanding with IUPAC Integration and Multi-task Tuning
Feb 27, 2024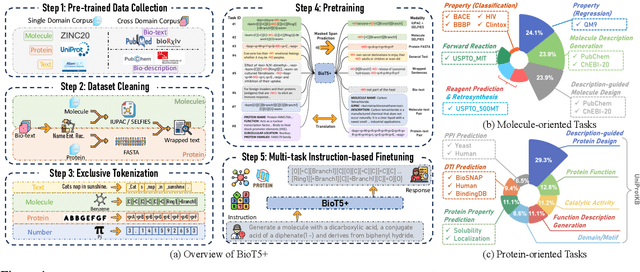
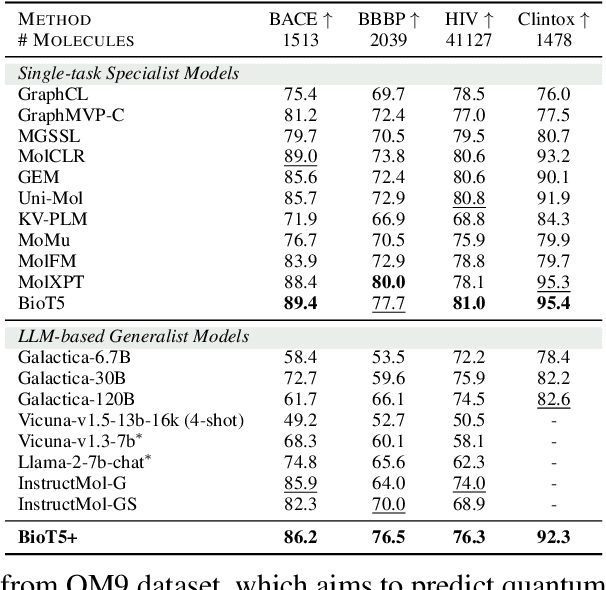
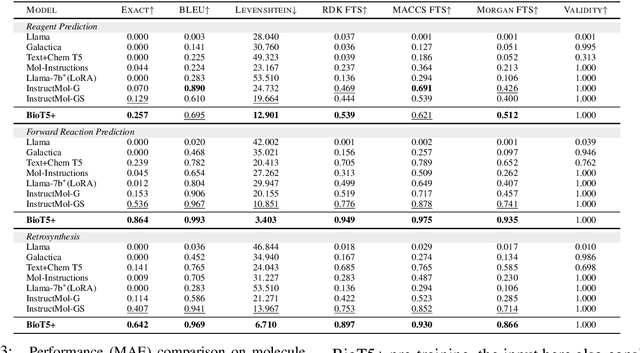
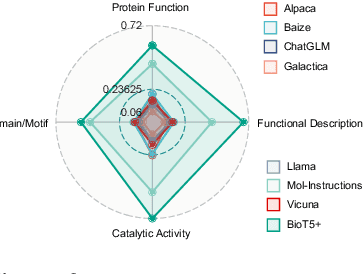
Abstract:Recent research trends in computational biology have increasingly focused on integrating text and bio-entity modeling, especially in the context of molecules and proteins. However, previous efforts like BioT5 faced challenges in generalizing across diverse tasks and lacked a nuanced understanding of molecular structures, particularly in their textual representations (e.g., IUPAC). This paper introduces BioT5+, an extension of the BioT5 framework, tailored to enhance biological research and drug discovery. BioT5+ incorporates several novel features: integration of IUPAC names for molecular understanding, inclusion of extensive bio-text and molecule data from sources like bioRxiv and PubChem, the multi-task instruction tuning for generality across tasks, and a novel numerical tokenization technique for improved processing of numerical data. These enhancements allow BioT5+ to bridge the gap between molecular representations and their textual descriptions, providing a more holistic understanding of biological entities, and largely improving the grounded reasoning of bio-text and bio-sequences. The model is pre-trained and fine-tuned with a large number of experiments, including \emph{3 types of problems (classification, regression, generation), 15 kinds of tasks, and 21 total benchmark datasets}, demonstrating the remarkable performance and state-of-the-art results in most cases. BioT5+ stands out for its ability to capture intricate relationships in biological data, thereby contributing significantly to bioinformatics and computational biology. Our code is available at \url{https://github.com/QizhiPei/BioT5}.
Knowledge Graphs Meet Multi-Modal Learning: A Comprehensive Survey
Feb 26, 2024



Abstract:Knowledge Graphs (KGs) play a pivotal role in advancing various AI applications, with the semantic web community's exploration into multi-modal dimensions unlocking new avenues for innovation. In this survey, we carefully review over 300 articles, focusing on KG-aware research in two principal aspects: KG-driven Multi-Modal (KG4MM) learning, where KGs support multi-modal tasks, and Multi-Modal Knowledge Graph (MM4KG), which extends KG studies into the MMKG realm. We begin by defining KGs and MMKGs, then explore their construction progress. Our review includes two primary task categories: KG-aware multi-modal learning tasks, such as Image Classification and Visual Question Answering, and intrinsic MMKG tasks like Multi-modal Knowledge Graph Completion and Entity Alignment, highlighting specific research trajectories. For most of these tasks, we provide definitions, evaluation benchmarks, and additionally outline essential insights for conducting relevant research. Finally, we discuss current challenges and identify emerging trends, such as progress in Large Language Modeling and Multi-modal Pre-training strategies. This survey aims to serve as a comprehensive reference for researchers already involved in or considering delving into KG and multi-modal learning research, offering insights into the evolving landscape of MMKG research and supporting future work.
ChatCell: Facilitating Single-Cell Analysis with Natural Language
Feb 20, 2024



Abstract:As Large Language Models (LLMs) rapidly evolve, their influence in science is becoming increasingly prominent. The emerging capabilities of LLMs in task generalization and free-form dialogue can significantly advance fields like chemistry and biology. However, the field of single-cell biology, which forms the foundational building blocks of living organisms, still faces several challenges. High knowledge barriers and limited scalability in current methods restrict the full exploitation of LLMs in mastering single-cell data, impeding direct accessibility and rapid iteration. To this end, we introduce ChatCell, which signifies a paradigm shift by facilitating single-cell analysis with natural language. Leveraging vocabulary adaptation and unified sequence generation, ChatCell has acquired profound expertise in single-cell biology and the capability to accommodate a diverse range of analysis tasks. Extensive experiments further demonstrate ChatCell's robust performance and potential to deepen single-cell insights, paving the way for more accessible and intuitive exploration in this pivotal field. Our project homepage is available at https://zjunlp.github.io/project/ChatCell.
Knowledgeable Preference Alignment for LLMs in Domain-specific Question Answering
Nov 11, 2023



Abstract:Recently, the development of large language models (LLMs) has attracted wide attention in academia and industry. Deploying LLMs to real scenarios is one of the key directions in the current Internet industry. In this paper, we present a novel pipeline to apply LLMs for domain-specific question answering (QA) that incorporates domain knowledge graphs (KGs), addressing an important direction of LLM application. As a real-world application, the content generated by LLMs should be user-friendly to serve the customers. Additionally, the model needs to utilize domain knowledge properly to generate reliable answers. These two issues are the two major difficulties in the LLM application as vanilla fine-tuning can not adequately address them. We think both requirements can be unified as the model preference problem that needs to align with humans to achieve practical application. Thus, we introduce Knowledgeable Preference AlignmenT (KnowPAT), which constructs two kinds of preference set called style preference set and knowledge preference set respectively to tackle the two issues. Besides, we design a new alignment objective to align the LLM preference with human preference, aiming to train a better LLM for real-scenario domain-specific QA to generate reliable and user-friendly answers. Adequate experiments and comprehensive with 15 baseline methods demonstrate that our KnowPAT is an outperforming pipeline for real-scenario domain-specific QA with LLMs. Our code is open-source at https://github.com/zjukg/KnowPAT.
Rethinking Uncertainly Missing and Ambiguous Visual Modality in Multi-Modal Entity Alignment
Aug 01, 2023Abstract:As a crucial extension of entity alignment (EA), multi-modal entity alignment (MMEA) aims to identify identical entities across disparate knowledge graphs (KGs) by exploiting associated visual information. However, existing MMEA approaches primarily concentrate on the fusion paradigm of multi-modal entity features, while neglecting the challenges presented by the pervasive phenomenon of missing and intrinsic ambiguity of visual images. In this paper, we present a further analysis of visual modality incompleteness, benchmarking latest MMEA models on our proposed dataset MMEA-UMVM, where the types of alignment KGs covering bilingual and monolingual, with standard (non-iterative) and iterative training paradigms to evaluate the model performance. Our research indicates that, in the face of modality incompleteness, models succumb to overfitting the modality noise, and exhibit performance oscillations or declines at high rates of missing modality. This proves that the inclusion of additional multi-modal data can sometimes adversely affect EA. To address these challenges, we introduce UMAEA , a robust multi-modal entity alignment approach designed to tackle uncertainly missing and ambiguous visual modalities. It consistently achieves SOTA performance across all 97 benchmark splits, significantly surpassing existing baselines with limited parameters and time consumption, while effectively alleviating the identified limitations of other models. Our code and benchmark data are available at https://github.com/zjukg/UMAEA.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge