Kangwei Liu
Where Not to Learn: Prior-Aligned Training with Subset-based Attribution Constraints for Reliable Decision-Making
Jan 30, 2026Abstract:Reliable models should not only predict correctly, but also justify decisions with acceptable evidence. Yet conventional supervised learning typically provides only class-level labels, allowing models to achieve high accuracy through shortcut correlations rather than the intended evidence. Human priors can help constrain such behavior, but aligning models to these priors remains challenging because learned representations often diverge from human perception. To address this challenge, we propose an attribution-based human prior alignment method. We encode human priors as input regions that the model is expected to rely on (e.g., bounding boxes), and leverage a highly faithful subset-selection-based attribution approach to expose the model's decision evidence during training. When the attribution region deviates substantially from the prior regions, we penalize reliance on off-prior evidence, encouraging the model to shift its attribution toward the intended regions. This is achieved through a training objective that imposes attribution constraints induced by the human prior. We validate our method on both image classification and click decision tasks in MLLM-based GUI agent models. Across conventional classification and autoregressive generation settings, human prior alignment consistently improves task accuracy while also enhancing the model's decision reasonability.
Where MLLMs Attend and What They Rely On: Explaining Autoregressive Token Generation
Sep 26, 2025Abstract:Multimodal large language models (MLLMs) have demonstrated remarkable capabilities in aligning visual inputs with natural language outputs. Yet, the extent to which generated tokens depend on visual modalities remains poorly understood, limiting interpretability and reliability. In this work, we present EAGLE, a lightweight black-box framework for explaining autoregressive token generation in MLLMs. EAGLE attributes any selected tokens to compact perceptual regions while quantifying the relative influence of language priors and perceptual evidence. The framework introduces an objective function that unifies sufficiency (insight score) and indispensability (necessity score), optimized via greedy search over sparsified image regions for faithful and efficient attribution. Beyond spatial attribution, EAGLE performs modality-aware analysis that disentangles what tokens rely on, providing fine-grained interpretability of model decisions. Extensive experiments across open-source MLLMs show that EAGLE consistently outperforms existing methods in faithfulness, localization, and hallucination diagnosis, while requiring substantially less GPU memory. These results highlight its effectiveness and practicality for advancing the interpretability of MLLMs. The code is available at https://github.com/RuoyuChen10/EAGLE.
ChineseHarm-Bench: A Chinese Harmful Content Detection Benchmark
Jun 12, 2025Abstract:Large language models (LLMs) have been increasingly applied to automated harmful content detection tasks, assisting moderators in identifying policy violations and improving the overall efficiency and accuracy of content review. However, existing resources for harmful content detection are predominantly focused on English, with Chinese datasets remaining scarce and often limited in scope. We present a comprehensive, professionally annotated benchmark for Chinese content harm detection, which covers six representative categories and is constructed entirely from real-world data. Our annotation process further yields a knowledge rule base that provides explicit expert knowledge to assist LLMs in Chinese harmful content detection. In addition, we propose a knowledge-augmented baseline that integrates both human-annotated knowledge rules and implicit knowledge from large language models, enabling smaller models to achieve performance comparable to state-of-the-art LLMs. Code and data are available at https://github.com/zjunlp/ChineseHarm-bench.
DisentTalk: Cross-lingual Talking Face Generation via Semantic Disentangled Diffusion Model
Mar 24, 2025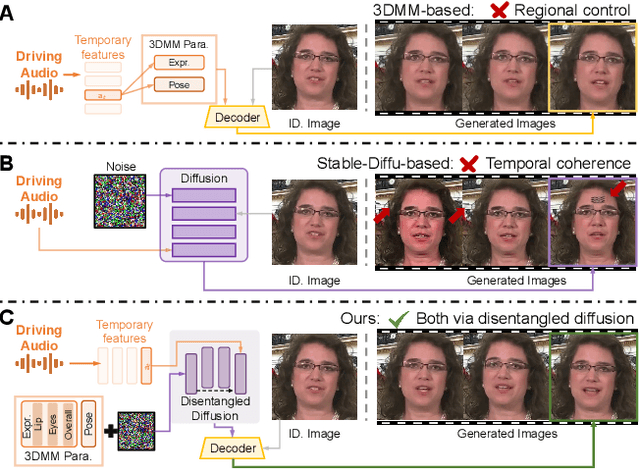
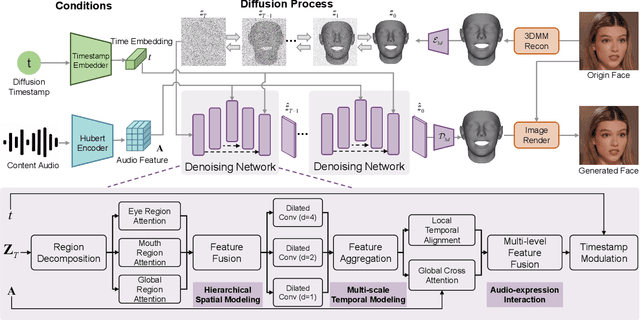
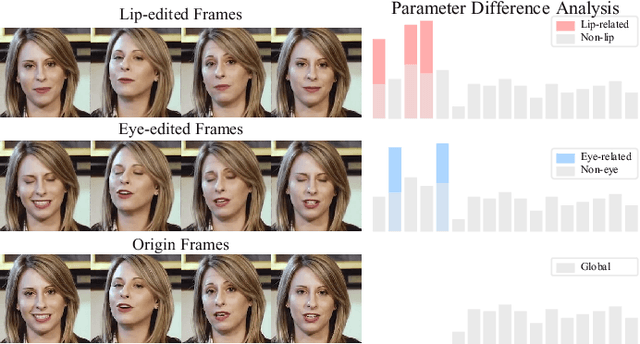

Abstract:Recent advances in talking face generation have significantly improved facial animation synthesis. However, existing approaches face fundamental limitations: 3DMM-based methods maintain temporal consistency but lack fine-grained regional control, while Stable Diffusion-based methods enable spatial manipulation but suffer from temporal inconsistencies. The integration of these approaches is hindered by incompatible control mechanisms and semantic entanglement of facial representations. This paper presents DisentTalk, introducing a data-driven semantic disentanglement framework that decomposes 3DMM expression parameters into meaningful subspaces for fine-grained facial control. Building upon this disentangled representation, we develop a hierarchical latent diffusion architecture that operates in 3DMM parameter space, integrating region-aware attention mechanisms to ensure both spatial precision and temporal coherence. To address the scarcity of high-quality Chinese training data, we introduce CHDTF, a Chinese high-definition talking face dataset. Extensive experiments show superior performance over existing methods across multiple metrics, including lip synchronization, expression quality, and temporal consistency. Project Page: https://kangweiiliu.github.io/DisentTalk.
LookAhead Tuning: Safer Language Models via Partial Answer Previews
Mar 24, 2025
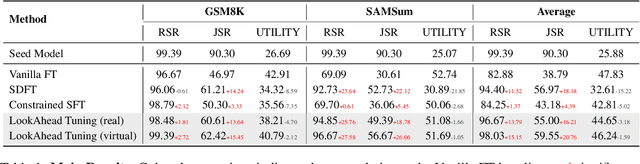
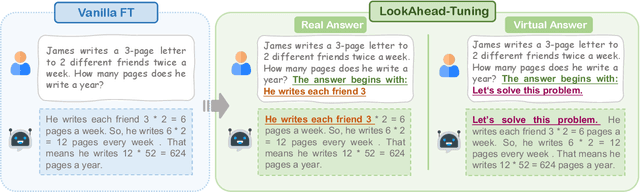
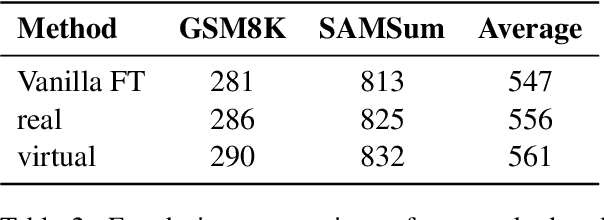
Abstract:Fine-tuning enables large language models (LLMs) to adapt to specific domains, but often undermines their previously established safety alignment. To mitigate the degradation of model safety during fine-tuning, we introduce LookAhead Tuning, which comprises two simple, low-resource, and effective data-driven methods that modify training data by previewing partial answer prefixes. Both methods aim to preserve the model's inherent safety mechanisms by minimizing perturbations to initial token distributions. Comprehensive experiments demonstrate that LookAhead Tuning effectively maintains model safety without sacrificing robust performance on downstream tasks. Our findings position LookAhead Tuning as a reliable and efficient solution for the safe and effective adaptation of LLMs. Code is released at https://github.com/zjunlp/LookAheadTuning.
A Multi-Modal AI Copilot for Single-Cell Analysis with Instruction Following
Jan 15, 2025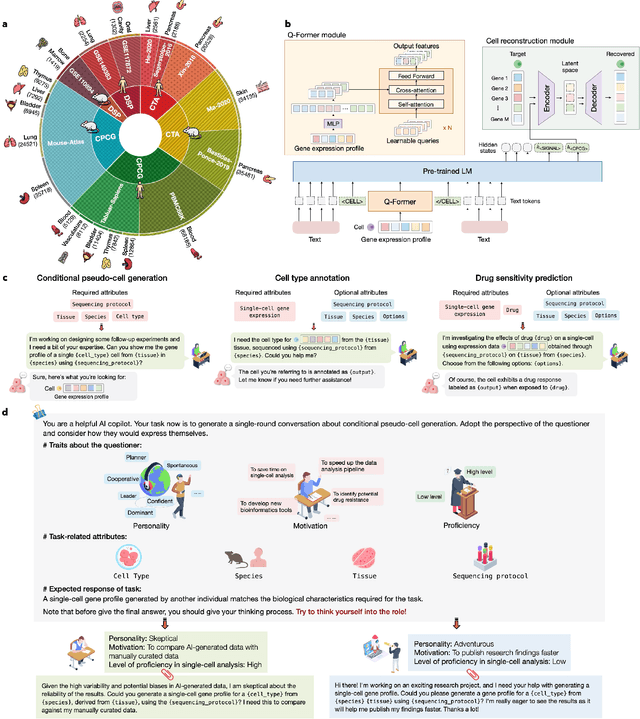
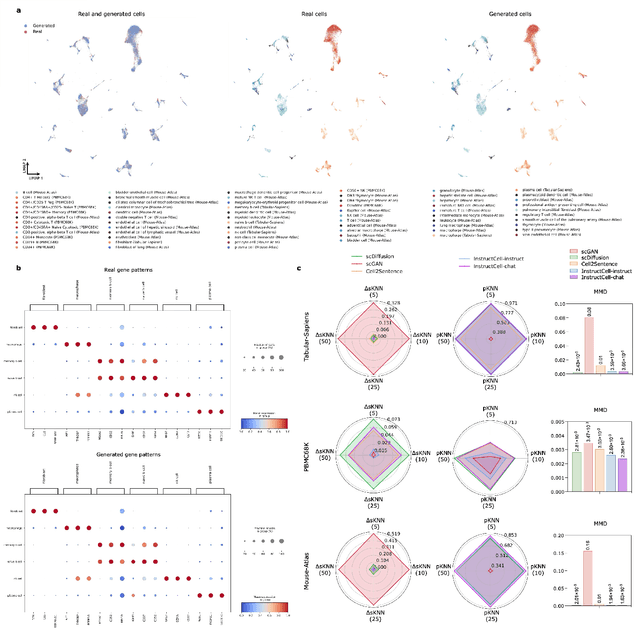
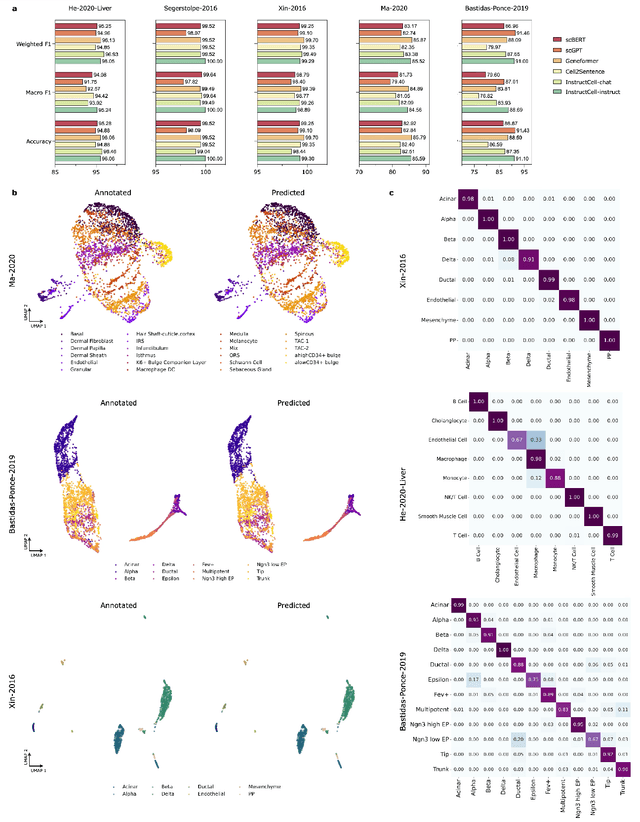
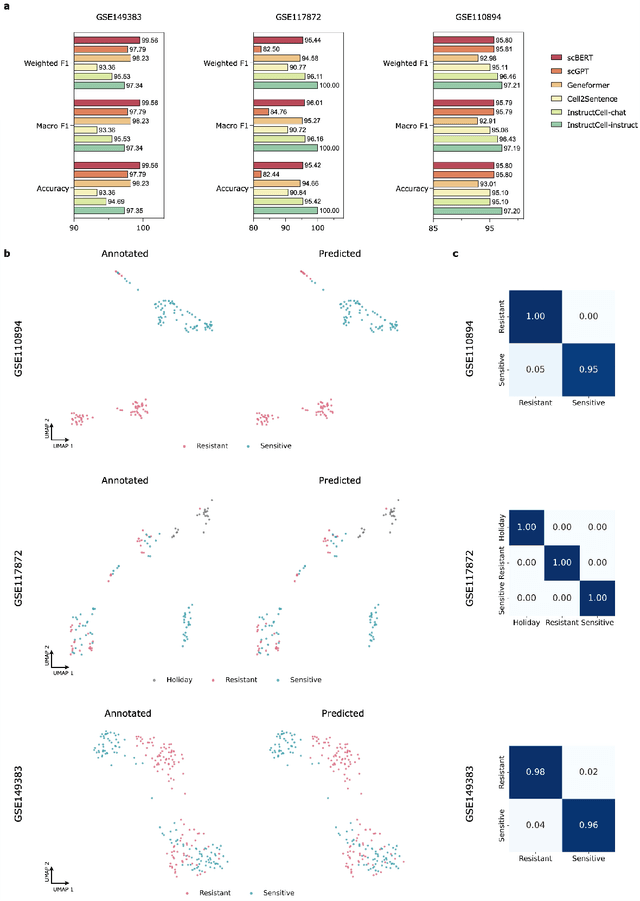
Abstract:Large language models excel at interpreting complex natural language instructions, enabling them to perform a wide range of tasks. In the life sciences, single-cell RNA sequencing (scRNA-seq) data serves as the "language of cellular biology", capturing intricate gene expression patterns at the single-cell level. However, interacting with this "language" through conventional tools is often inefficient and unintuitive, posing challenges for researchers. To address these limitations, we present InstructCell, a multi-modal AI copilot that leverages natural language as a medium for more direct and flexible single-cell analysis. We construct a comprehensive multi-modal instruction dataset that pairs text-based instructions with scRNA-seq profiles from diverse tissues and species. Building on this, we develop a multi-modal cell language architecture capable of simultaneously interpreting and processing both modalities. InstructCell empowers researchers to accomplish critical tasks-such as cell type annotation, conditional pseudo-cell generation, and drug sensitivity prediction-using straightforward natural language commands. Extensive evaluations demonstrate that InstructCell consistently meets or exceeds the performance of existing single-cell foundation models, while adapting to diverse experimental conditions. More importantly, InstructCell provides an accessible and intuitive tool for exploring complex single-cell data, lowering technical barriers and enabling deeper biological insights.
OneKE: A Dockerized Schema-Guided LLM Agent-based Knowledge Extraction System
Dec 28, 2024


Abstract:We introduce OneKE, a dockerized schema-guided knowledge extraction system, which can extract knowledge from the Web and raw PDF Books, and support various domains (science, news, etc.). Specifically, we design OneKE with multiple agents and a configure knowledge base. Different agents perform their respective roles, enabling support for various extraction scenarios. The configure knowledge base facilitates schema configuration, error case debugging and correction, further improving the performance. Empirical evaluations on benchmark datasets demonstrate OneKE's efficacy, while case studies further elucidate its adaptability to diverse tasks across multiple domains, highlighting its potential for broad applications. We have open-sourced the Code at https://github.com/zjunlp/OneKE and released a Video at http://oneke.openkg.cn/demo.mp4.
ChatCell: Facilitating Single-Cell Analysis with Natural Language
Feb 20, 2024



Abstract:As Large Language Models (LLMs) rapidly evolve, their influence in science is becoming increasingly prominent. The emerging capabilities of LLMs in task generalization and free-form dialogue can significantly advance fields like chemistry and biology. However, the field of single-cell biology, which forms the foundational building blocks of living organisms, still faces several challenges. High knowledge barriers and limited scalability in current methods restrict the full exploitation of LLMs in mastering single-cell data, impeding direct accessibility and rapid iteration. To this end, we introduce ChatCell, which signifies a paradigm shift by facilitating single-cell analysis with natural language. Leveraging vocabulary adaptation and unified sequence generation, ChatCell has acquired profound expertise in single-cell biology and the capability to accommodate a diverse range of analysis tasks. Extensive experiments further demonstrate ChatCell's robust performance and potential to deepen single-cell insights, paving the way for more accessible and intuitive exploration in this pivotal field. Our project homepage is available at https://zjunlp.github.io/project/ChatCell.
EasyInstruct: An Easy-to-use Instruction Processing Framework for Large Language Models
Feb 06, 2024



Abstract:In recent years, instruction tuning has gained increasing attention and emerged as a crucial technique to enhance the capabilities of Large Language Models (LLMs). To construct high-quality instruction datasets, many instruction processing approaches have been proposed, aiming to achieve a delicate balance between data quantity and data quality. Nevertheless, due to inconsistencies that persist among various instruction processing methods, there is no standard open-source instruction processing implementation framework available for the community, which hinders practitioners from further developing and advancing. To facilitate instruction processing research and development, we present EasyInstruct, an easy-to-use instruction processing framework for LLMs, which modularizes instruction generation, selection, and prompting, while also considering their combination and interaction. EasyInstruct is publicly released and actively maintained at https://github.com/zjunlp/EasyInstruct, along with a running demo App at https://huggingface.co/spaces/zjunlp/EasyInstruct for quick-start, calling for broader research centered on instruction data.
EasyEdit: An Easy-to-use Knowledge Editing Framework for Large Language Models
Aug 14, 2023Abstract:Large Language Models (LLMs) usually suffer from knowledge cutoff or fallacy issues, which means they are unaware of unseen events or generate text with incorrect facts owing to the outdated/noisy data. To this end, many knowledge editing approaches for LLMs have emerged -- aiming to subtly inject/edit updated knowledge or adjust undesired behavior while minimizing the impact on unrelated inputs. Nevertheless, due to significant differences among various knowledge editing methods and the variations in task setups, there is no standard implementation framework available for the community, which hinders practitioners to apply knowledge editing to applications. To address these issues, we propose EasyEdit, an easy-to-use knowledge editing framework for LLMs. It supports various cutting-edge knowledge editing approaches and can be readily apply to many well-known LLMs such as T5, GPT-J, LlaMA, etc. Empirically, we report the knowledge editing results on LlaMA-2 with EasyEdit, demonstrating that knowledge editing surpasses traditional fine-tuning in terms of reliability and generalization. We have released the source code on GitHub at https://github.com/zjunlp/EasyEdit, along with Google Colab tutorials and comprehensive documentation for beginners to get started. Besides, we present an online system for real-time knowledge editing, and a demo video at http://knowlm.zjukg.cn/easyedit.mp4.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge