Xiaozhuan Liang
ChineseHarm-Bench: A Chinese Harmful Content Detection Benchmark
Jun 12, 2025Abstract:Large language models (LLMs) have been increasingly applied to automated harmful content detection tasks, assisting moderators in identifying policy violations and improving the overall efficiency and accuracy of content review. However, existing resources for harmful content detection are predominantly focused on English, with Chinese datasets remaining scarce and often limited in scope. We present a comprehensive, professionally annotated benchmark for Chinese content harm detection, which covers six representative categories and is constructed entirely from real-world data. Our annotation process further yields a knowledge rule base that provides explicit expert knowledge to assist LLMs in Chinese harmful content detection. In addition, we propose a knowledge-augmented baseline that integrates both human-annotated knowledge rules and implicit knowledge from large language models, enabling smaller models to achieve performance comparable to state-of-the-art LLMs. Code and data are available at https://github.com/zjunlp/ChineseHarm-bench.
SFM-Protein: Integrative Co-evolutionary Pre-training for Advanced Protein Sequence Representation
Oct 31, 2024



Abstract:Proteins, essential to biological systems, perform functions intricately linked to their three-dimensional structures. Understanding the relationship between protein structures and their amino acid sequences remains a core challenge in protein modeling. While traditional protein foundation models benefit from pre-training on vast unlabeled datasets, they often struggle to capture critical co-evolutionary information, which evolutionary-based methods excel at. In this study, we introduce a novel pre-training strategy for protein foundation models that emphasizes the interactions among amino acid residues to enhance the extraction of both short-range and long-range co-evolutionary features from sequence data. Trained on a large-scale protein sequence dataset, our model demonstrates superior generalization ability, outperforming established baselines of similar size, including the ESM model, across diverse downstream tasks. Experimental results confirm the model's effectiveness in integrating co-evolutionary information, marking a significant step forward in protein sequence-based modeling.
To Forget or Not? Towards Practical Knowledge Unlearning for Large Language Models
Jul 02, 2024



Abstract:Large Language Models (LLMs) trained on extensive corpora inevitably retain sensitive data, such as personal privacy information and copyrighted material. Recent advancements in knowledge unlearning involve updating LLM parameters to erase specific knowledge. However, current unlearning paradigms are mired in vague forgetting boundaries, often erasing knowledge indiscriminately. In this work, we introduce KnowUnDo, a benchmark containing copyrighted content and user privacy domains to evaluate if the unlearning process inadvertently erases essential knowledge. Our findings indicate that existing unlearning methods often suffer from excessive unlearning. To address this, we propose a simple yet effective method, MemFlex, which utilizes gradient information to precisely target and unlearn sensitive parameters. Experimental results show that MemFlex is superior to existing methods in both precise knowledge unlearning and general knowledge retaining of LLMs. Code and dataset will be released at https://github.com/zjunlp/KnowUnDo.
BioT5+: Towards Generalized Biological Understanding with IUPAC Integration and Multi-task Tuning
Feb 27, 2024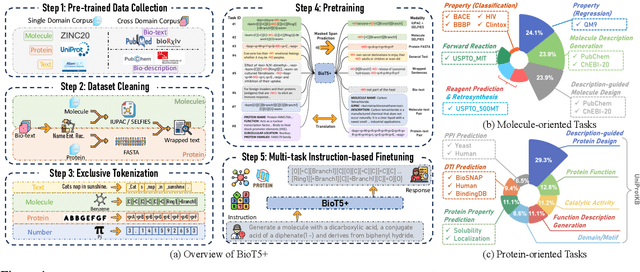
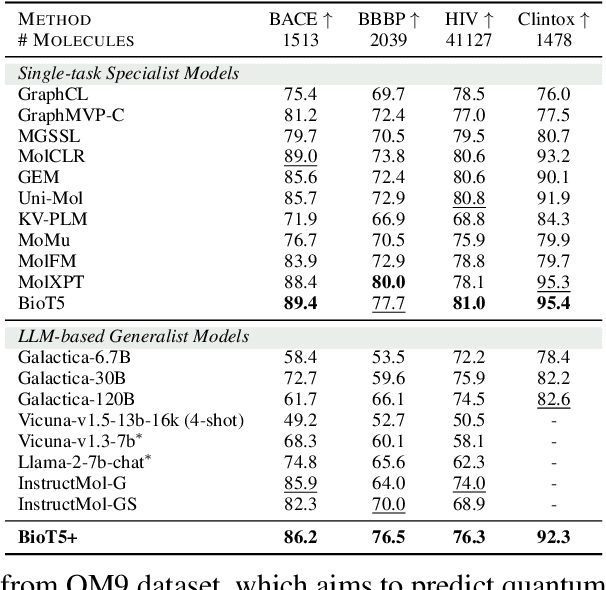
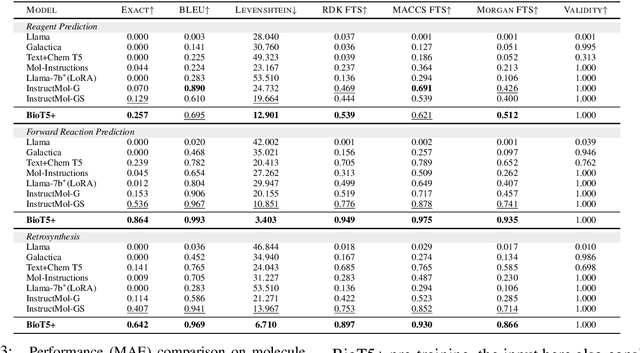
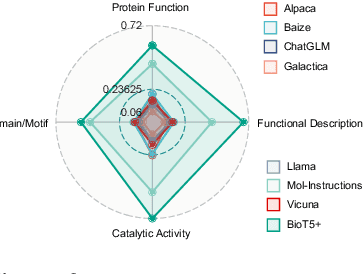
Abstract:Recent research trends in computational biology have increasingly focused on integrating text and bio-entity modeling, especially in the context of molecules and proteins. However, previous efforts like BioT5 faced challenges in generalizing across diverse tasks and lacked a nuanced understanding of molecular structures, particularly in their textual representations (e.g., IUPAC). This paper introduces BioT5+, an extension of the BioT5 framework, tailored to enhance biological research and drug discovery. BioT5+ incorporates several novel features: integration of IUPAC names for molecular understanding, inclusion of extensive bio-text and molecule data from sources like bioRxiv and PubChem, the multi-task instruction tuning for generality across tasks, and a novel numerical tokenization technique for improved processing of numerical data. These enhancements allow BioT5+ to bridge the gap between molecular representations and their textual descriptions, providing a more holistic understanding of biological entities, and largely improving the grounded reasoning of bio-text and bio-sequences. The model is pre-trained and fine-tuned with a large number of experiments, including \emph{3 types of problems (classification, regression, generation), 15 kinds of tasks, and 21 total benchmark datasets}, demonstrating the remarkable performance and state-of-the-art results in most cases. BioT5+ stands out for its ability to capture intricate relationships in biological data, thereby contributing significantly to bioinformatics and computational biology. Our code is available at \url{https://github.com/QizhiPei/BioT5}.
InstructEdit: Instruction-based Knowledge Editing for Large Language Models
Feb 25, 2024



Abstract:Knowledge editing for large language models can offer an efficient solution to alter a model's behavior without negatively impacting the overall performance. However, the current approach encounters issues with limited generalizability across tasks, necessitating one distinct editor for each task, which significantly hinders the broader applications. To address this, we take the first step to analyze the multi-task generalization issue in knowledge editing. Specifically, we develop an instruction-based editing technique, termed InstructEdit, which facilitates the editor's adaptation to various task performances simultaneously using simple instructions. With only one unified editor for each LLM, we empirically demonstrate that InstructEdit can improve the editor's control, leading to an average 14.86% increase in Reliability in multi-task editing setting. Furthermore, experiments involving holdout unseen task illustrate that InstructEdit consistently surpass previous strong baselines. To further investigate the underlying mechanisms of instruction-based knowledge editing, we analyze the principal components of the editing gradient directions, which unveils that instructions can help control optimization direction with stronger OOD generalization. Code and datasets will be available in https://github.com/zjunlp/EasyEdit.
Mol-Instructions: A Large-Scale Biomolecular Instruction Dataset for Large Language Models
Jun 13, 2023


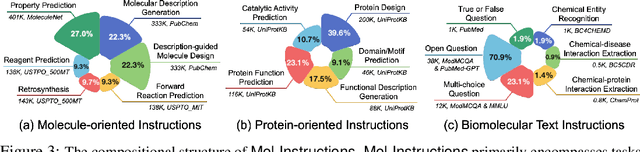
Abstract:Large Language Models (LLMs), with their remarkable task-handling capabilities and innovative outputs, have catalyzed significant advancements across a spectrum of fields. However, their proficiency within specialized domains such as biomolecular studies remains limited. To address this challenge, we introduce Mol-Instructions, a meticulously curated, comprehensive instruction dataset expressly designed for the biomolecular realm. Mol-Instructions is composed of three pivotal components: molecule-oriented instructions, protein-oriented instructions, and biomolecular text instructions, each curated to enhance the understanding and prediction capabilities of LLMs concerning biomolecular features and behaviors. Through extensive instruction tuning experiments on the representative LLM, we underscore the potency of Mol-Instructions to enhance the adaptability and cognitive acuity of large models within the complex sphere of biomolecular studies, thereby promoting advancements in the biomolecular research community. Mol-Instructions is made publicly accessible for future research endeavors and will be subjected to continual updates for enhanced applicability.
Multimodal Analogical Reasoning over Knowledge Graphs
Oct 01, 2022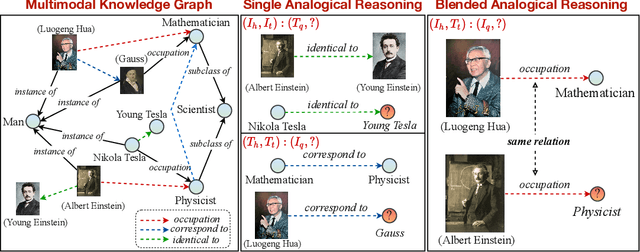

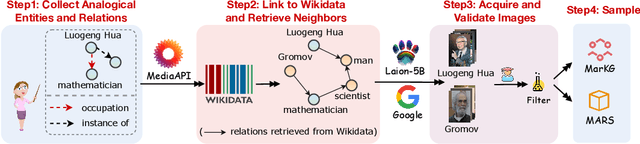
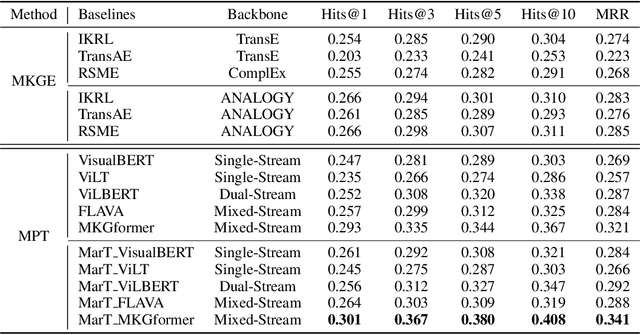
Abstract:Analogical reasoning is fundamental to human cognition and holds an important place in various fields. However, previous studies mainly focus on single-modal analogical reasoning and ignore taking advantage of structure knowledge. Notably, the research in cognitive psychology has demonstrated that information from multimodal sources always brings more powerful cognitive transfer than single modality sources. To this end, we introduce the new task of multimodal analogical reasoning over knowledge graphs, which requires multimodal reasoning ability with the help of background knowledge. Specifically, we construct a Multimodal Analogical Reasoning dataSet (MARS) and a multimodal knowledge graph MarKG. We evaluate with multimodal knowledge graph embedding and pre-trained Transformer baselines, illustrating the potential challenges of the proposed task. We further propose a novel model-agnostic Multimodal analogical reasoning framework with Transformer (MarT) motivated by the structure mapping theory, which can obtain better performance.
Decoupling Knowledge from Memorization: Retrieval-augmented Prompt Learning
May 29, 2022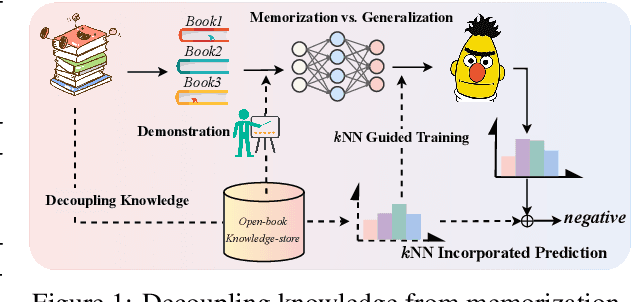
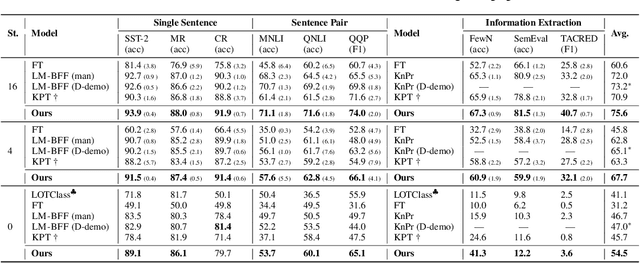
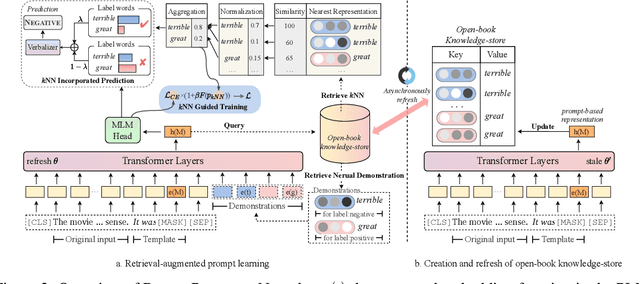
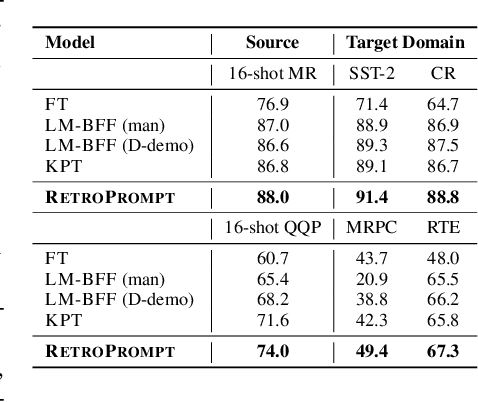
Abstract:Prompt learning approaches have made waves in natural language processing by inducing better few-shot performance while they still follow a parametric-based learning paradigm; the oblivion and rote memorization problems in learning may encounter unstable generalization issues. Specifically, vanilla prompt learning may struggle to utilize atypical instances by rote during fully-supervised training or overfit shallow patterns with low-shot data. To alleviate such limitations, we develop RetroPrompt with the motivation of decoupling knowledge from memorization to help the model strike a balance between generalization and memorization. In contrast with vanilla prompt learning, RetroPrompt constructs an open-book knowledge-store from training instances and implements a retrieval mechanism during the process of input, training and inference, thus equipping the model with the ability to retrieve related contexts from the training corpus as cues for enhancement. Extensive experiments demonstrate that RetroPrompt can obtain better performance in both few-shot and zero-shot settings. Besides, we further illustrate that our proposed RetroPrompt can yield better generalization abilities with new datasets. Detailed analysis of memorization indeed reveals RetroPrompt can reduce the reliance of language models on memorization; thus, improving generalization for downstream tasks.
Relphormer: Relational Graph Transformer for Knowledge Graph Representation
May 24, 2022
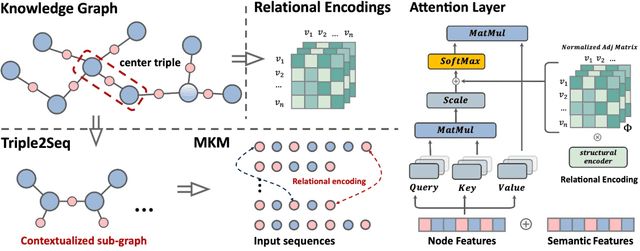
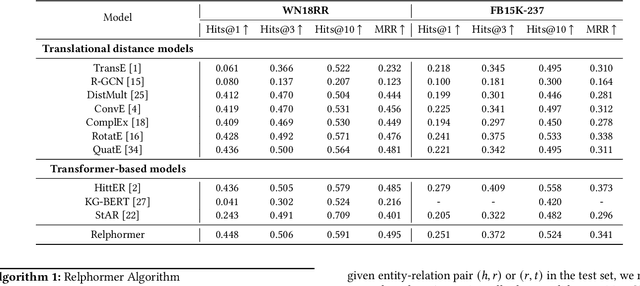
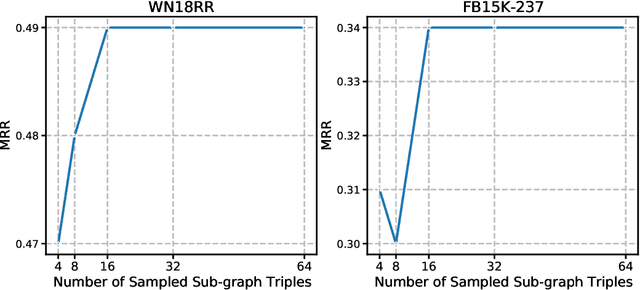
Abstract:Transformers have achieved remarkable performance in widespread fields, including natural language processing, computer vision and graph mining. However, in the knowledge graph representation, where translational distance paradigm dominates this area, vanilla Transformer architectures have not yielded promising improvements. Note that vanilla Transformer architectures struggle to capture the intrinsically semantic and structural information of knowledge graphs and can hardly scale to long-distance neighbors due to quadratic dependency. To this end, we propose a new variant of Transformer for knowledge graph representation dubbed Relphormer. Specifically, we introduce Triple2Seq which can dynamically sample contextualized sub-graph sequences as the input of the Transformer to alleviate the scalability issue. We then propose a novel structure-enhanced self-attention mechanism to encode the relational information and keep the globally semantic information among sub-graphs. Moreover, we propose masked knowledge modeling as a new paradigm for knowledge graph representation learning to unify different link prediction tasks. Experimental results show that our approach can obtain better performance on benchmark datasets compared with baselines.
Contrastive Demonstration Tuning for Pre-trained Language Models
Apr 18, 2022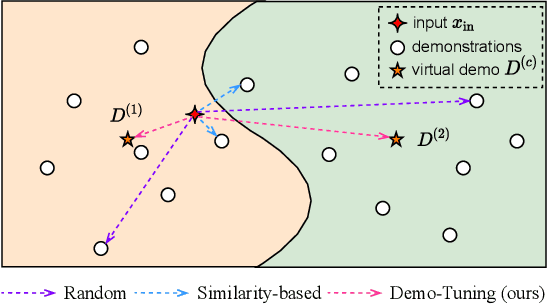
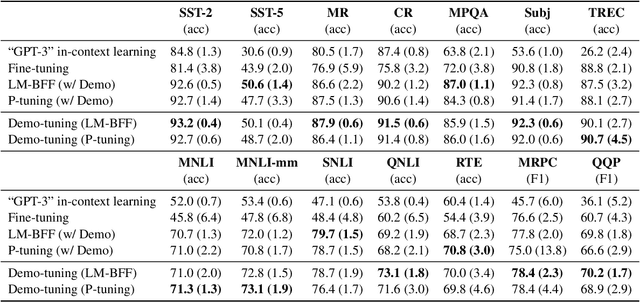
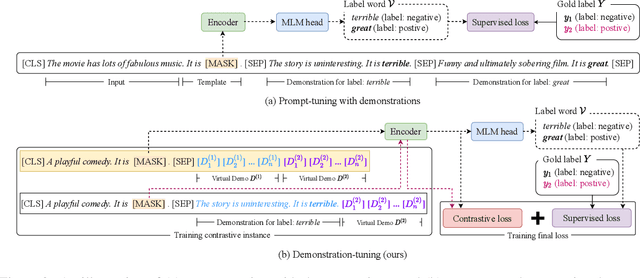
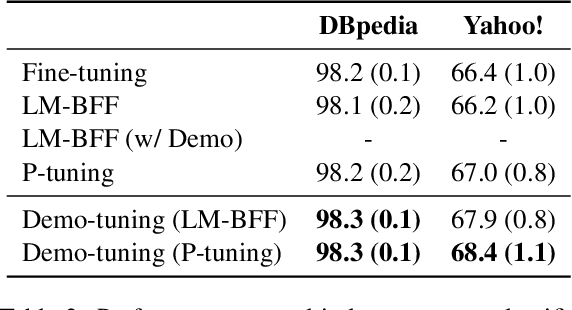
Abstract:Pretrained language models can be effectively stimulated by textual prompts or demonstrations, especially in low-data scenarios. Recent works have focused on automatically searching discrete or continuous prompts or optimized verbalizers, yet studies for the demonstration are still limited. Concretely, the demonstration examples are crucial for an excellent final performance of prompt-tuning. In this paper, we propose a novel pluggable, extensible, and efficient approach named contrastive demonstration tuning, which is free of demonstration sampling. Furthermore, the proposed approach can be: (i) Plugged to any previous prompt-tuning approaches; (ii) Extended to widespread classification tasks with a large number of categories. Experimental results on 16 datasets illustrate that our method integrated with previous approaches LM-BFF and P-tuning can yield better performance. Code is available in https://github.com/zjunlp/PromptKG/tree/main/research/Demo-Tuning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge