Penghui Yang
MATAI: A Generalist Machine Learning Framework for Property Prediction and Inverse Design of Advanced Alloys
Nov 13, 2025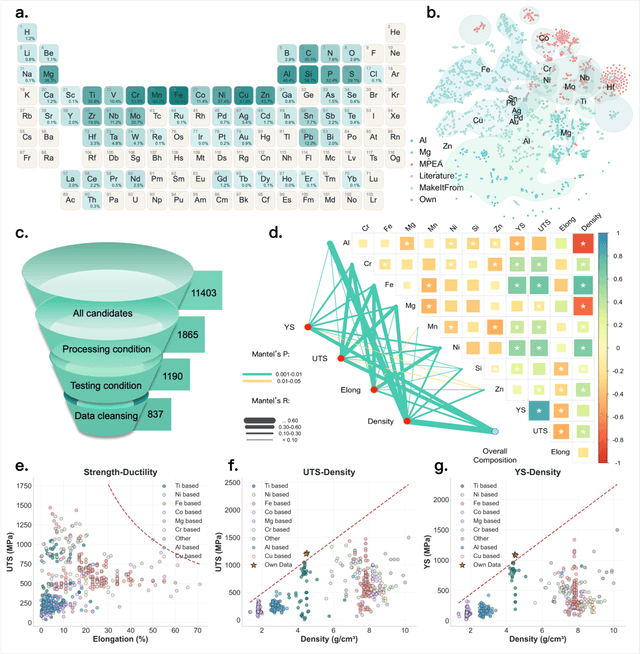
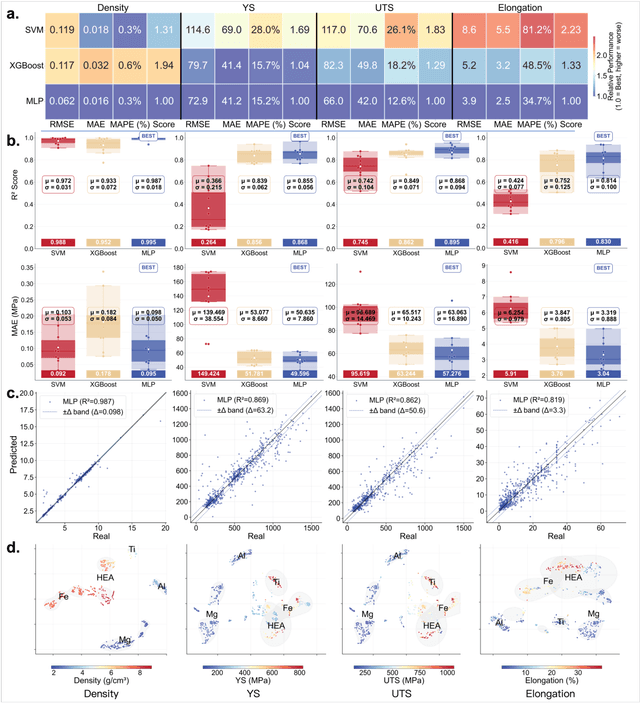
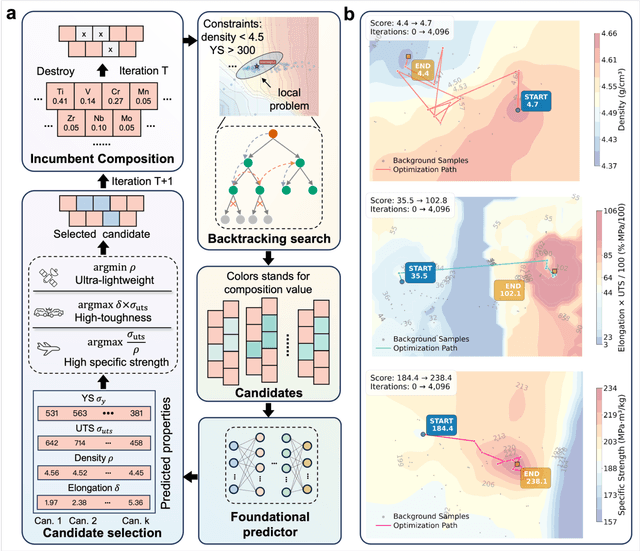
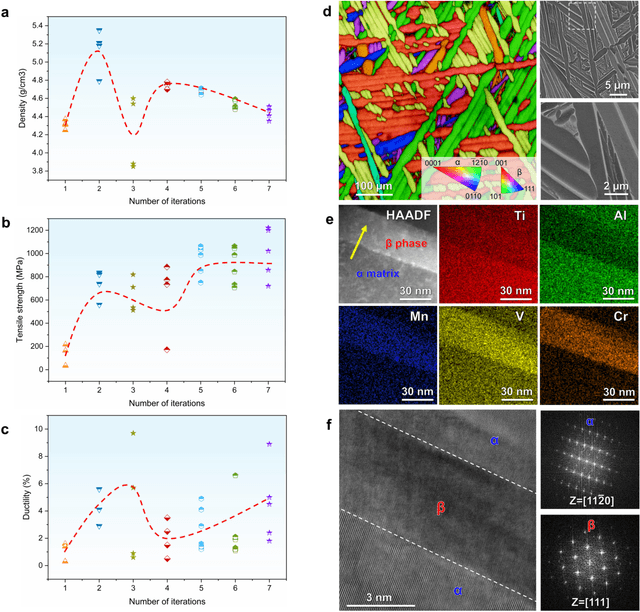
Abstract:The discovery of advanced metallic alloys is hindered by vast composition spaces, competing property objectives, and real-world constraints on manufacturability. Here we introduce MATAI, a generalist machine learning framework for property prediction and inverse design of as-cast alloys. MATAI integrates a curated alloy database, deep neural network-based property predictors, a constraint-aware optimization engine, and an iterative AI-experiment feedback loop. The framework estimates key mechanical propertie, sincluding density, yield strength, ultimate tensile strength, and elongation, directly from composition, using multi-task learning and physics-informed inductive biases. Alloy design is framed as a constrained optimization problem and solved using a bi-level approach that combines local search with symbolic constraint programming. We demonstrate MATAI's capabilities on the Ti-based alloy system, a canonical class of lightweight structural materials, where it rapidly identifies candidates that simultaneously achieve lower density (<4.45 g/cm3), higher strength (>1000 MPa) and appreciable ductility (>5%) through only seven iterations. Experimental validation confirms that MATAI-designed alloys outperform commercial references such as TC4, highlighting the framework's potential to accelerate the discovery of lightweight, high-performance materials under real-world design constraints.
OpenFly: A Versatile Toolchain and Large-scale Benchmark for Aerial Vision-Language Navigation
Feb 25, 2025Abstract:Vision-Language Navigation (VLN) aims to guide agents through an environment by leveraging both language instructions and visual cues, playing a pivotal role in embodied AI. Indoor VLN has been extensively studied, whereas outdoor aerial VLN remains underexplored. The potential reason is that outdoor aerial view encompasses vast areas, making data collection more challenging, which results in a lack of benchmarks. To address this problem, we propose OpenFly, a platform comprising a versatile toolchain and large-scale benchmark for aerial VLN. Firstly, we develop a highly automated toolchain for data collection, enabling automatic point cloud acquisition, scene semantic segmentation, flight trajectory creation, and instruction generation. Secondly, based on the toolchain, we construct a large-scale aerial VLN dataset with 100k trajectories, covering diverse heights and lengths across 18 scenes. The corresponding visual data are generated using various rendering engines and advanced techniques, including Unreal Engine, GTA V, Google Earth, and 3D Gaussian Splatting (3D GS). All data exhibit high visual quality. Particularly, 3D GS supports real-to-sim rendering, further enhancing the realism of the dataset. Thirdly, we propose OpenFly-Agent, a keyframe-aware VLN model, which takes language instructions, current observations, and historical keyframes as input, and outputs flight actions directly. Extensive analyses and experiments are conducted, showcasing the superiority of our OpenFly platform and OpenFly-Agent. The toolchain, dataset, and codes will be open-sourced.
LongSpec: Long-Context Speculative Decoding with Efficient Drafting and Verification
Feb 24, 2025
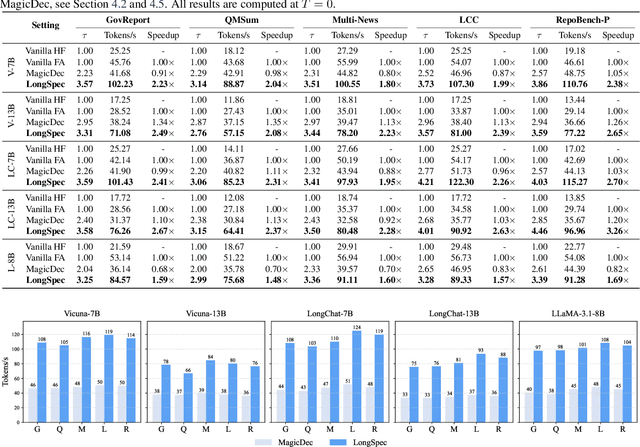
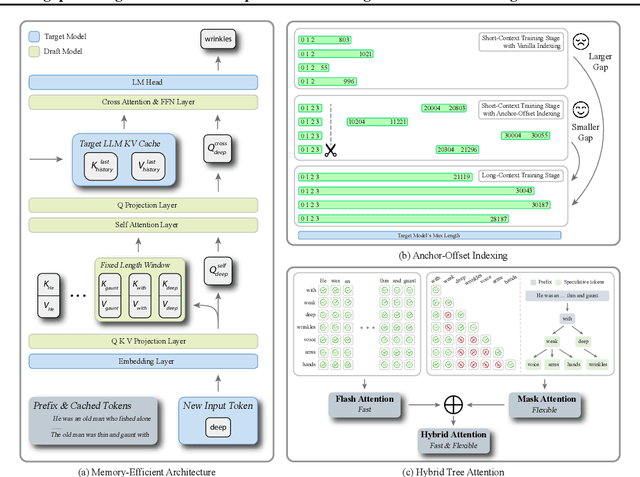
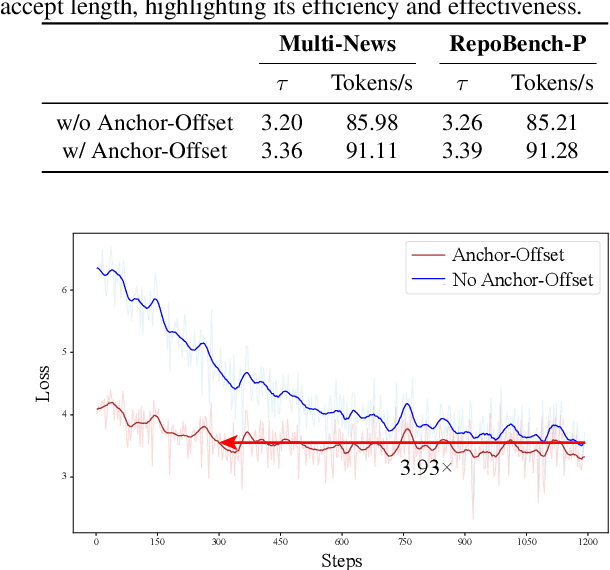
Abstract:Speculative decoding has become a promising technique to mitigate the high inference latency of autoregressive decoding in Large Language Models (LLMs). Despite its promise, the effective application of speculative decoding in LLMs still confronts three key challenges: the increasing memory demands of the draft model, the distribution shift between the short-training corpora and long-context inference, and inefficiencies in attention implementation. In this work, we enhance the performance of speculative decoding in long-context settings by addressing these challenges. First, we propose a memory-efficient draft model with a constant-sized Key-Value (KV) cache. Second, we introduce novel position indices for short-training data, enabling seamless adaptation from short-context training to long-context inference. Finally, we present an innovative attention aggregation method that combines fast implementations for prefix computation with standard attention for tree mask handling, effectively resolving the latency and memory inefficiencies of tree decoding. Our approach achieves strong results on various long-context tasks, including repository-level code completion, long-context summarization, and o1-like long reasoning tasks, demonstrating significant improvements in latency reduction. The code is available at https://github.com/sail-sg/LongSpec.
Sailor2: Sailing in South-East Asia with Inclusive Multilingual LLMs
Feb 18, 2025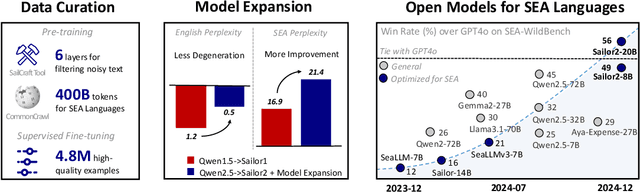
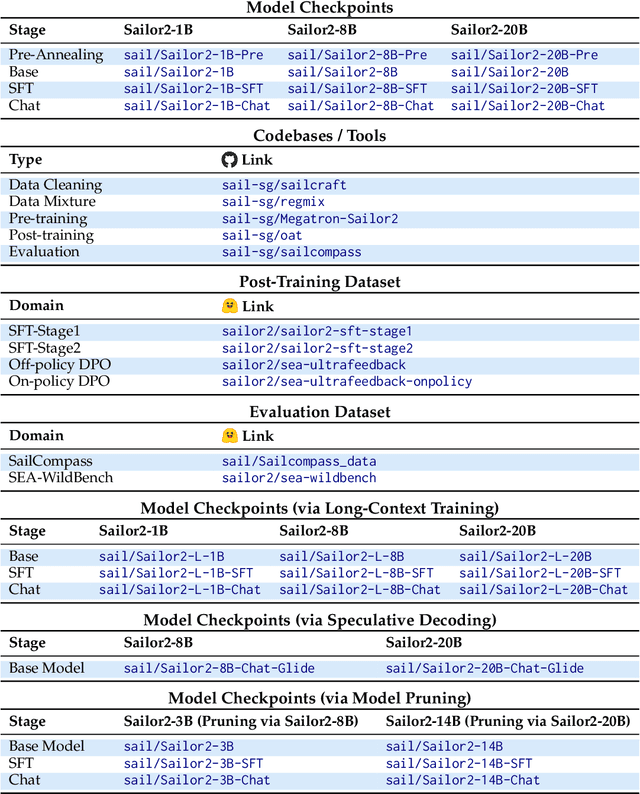
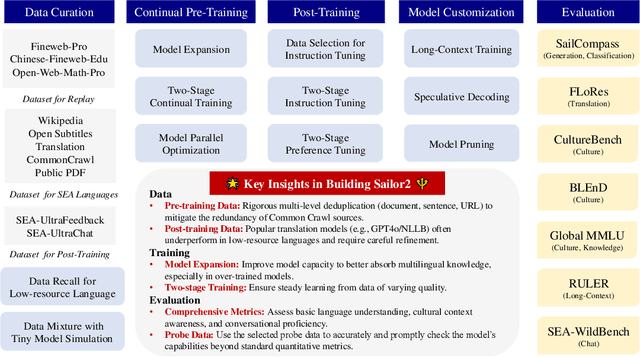
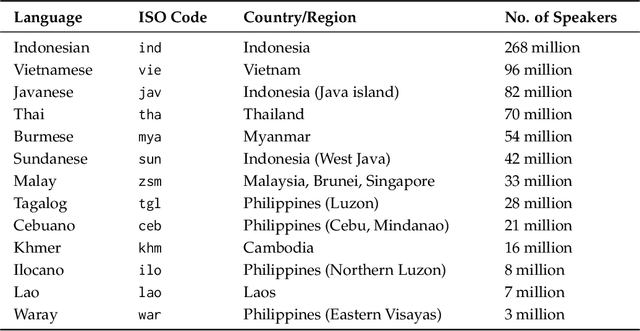
Abstract:Sailor2 is a family of cutting-edge multilingual language models for South-East Asian (SEA) languages, available in 1B, 8B, and 20B sizes to suit diverse applications. Building on Qwen2.5, Sailor2 undergoes continuous pre-training on 500B tokens (400B SEA-specific and 100B replay tokens) to support 13 SEA languages while retaining proficiency in Chinese and English. Sailor2-20B model achieves a 50-50 win rate against GPT-4o across SEA languages. We also deliver a comprehensive cookbook on how to develop the multilingual model in an efficient manner, including five key aspects: data curation, pre-training, post-training, model customization and evaluation. We hope that Sailor2 model (Apache 2.0 license) will drive language development in the SEA region, and Sailor2 cookbook will inspire researchers to build more inclusive LLMs for other under-served languages.
A Multi-Modal AI Copilot for Single-Cell Analysis with Instruction Following
Jan 15, 2025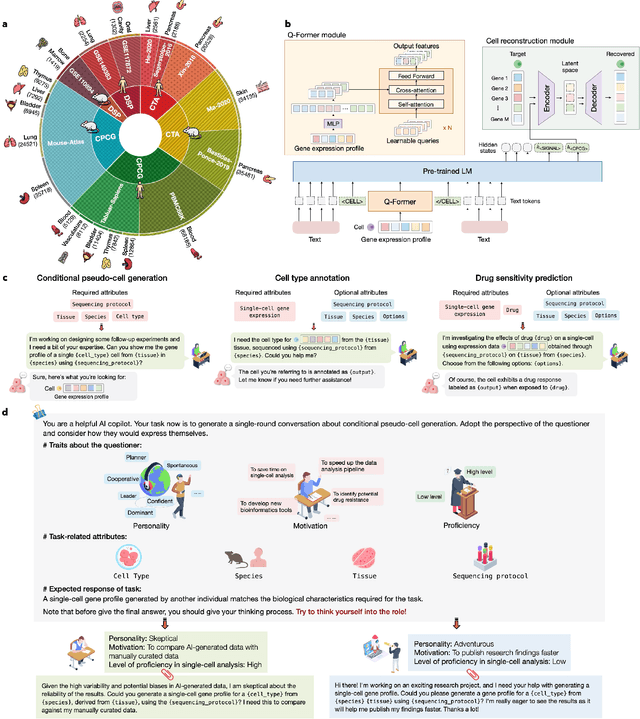
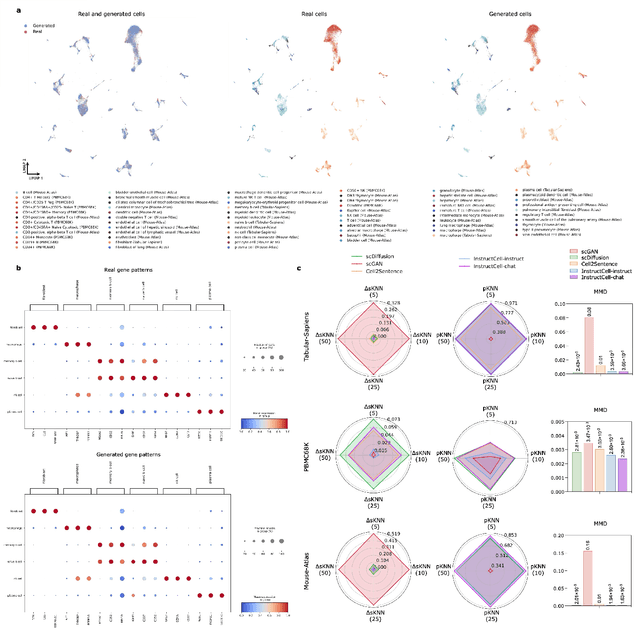
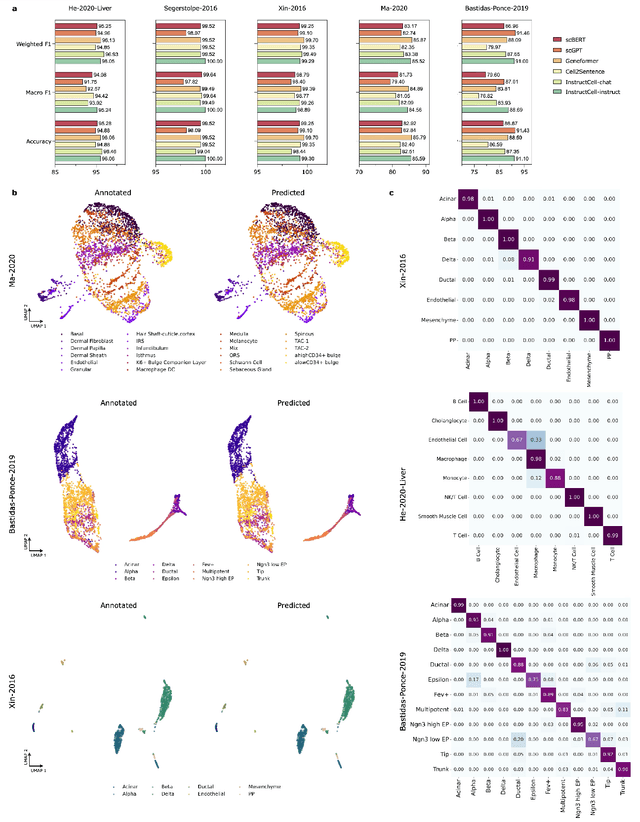
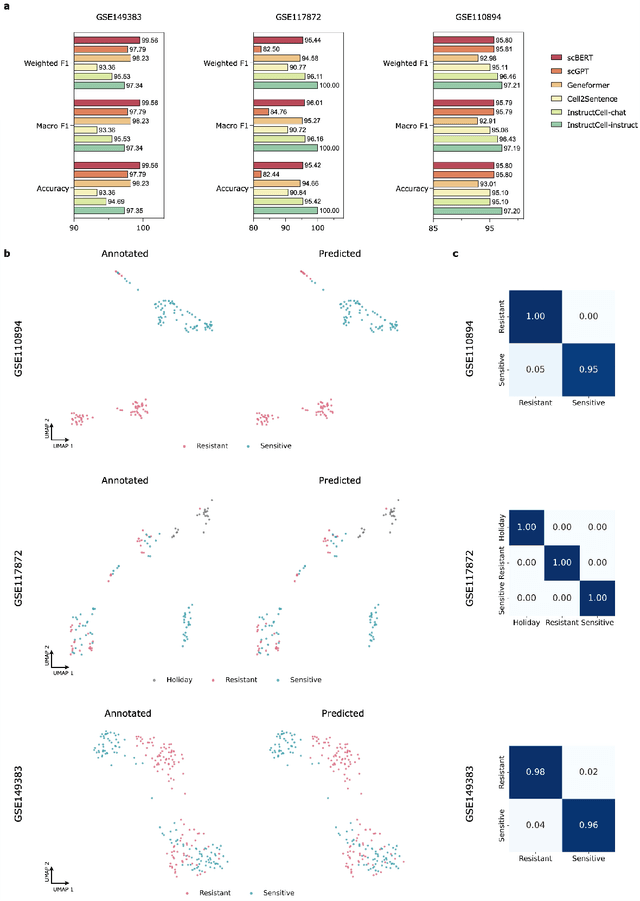
Abstract:Large language models excel at interpreting complex natural language instructions, enabling them to perform a wide range of tasks. In the life sciences, single-cell RNA sequencing (scRNA-seq) data serves as the "language of cellular biology", capturing intricate gene expression patterns at the single-cell level. However, interacting with this "language" through conventional tools is often inefficient and unintuitive, posing challenges for researchers. To address these limitations, we present InstructCell, a multi-modal AI copilot that leverages natural language as a medium for more direct and flexible single-cell analysis. We construct a comprehensive multi-modal instruction dataset that pairs text-based instructions with scRNA-seq profiles from diverse tissues and species. Building on this, we develop a multi-modal cell language architecture capable of simultaneously interpreting and processing both modalities. InstructCell empowers researchers to accomplish critical tasks-such as cell type annotation, conditional pseudo-cell generation, and drug sensitivity prediction-using straightforward natural language commands. Extensive evaluations demonstrate that InstructCell consistently meets or exceeds the performance of existing single-cell foundation models, while adapting to diverse experimental conditions. More importantly, InstructCell provides an accessible and intuitive tool for exploring complex single-cell data, lowering technical barriers and enabling deeper biological insights.
Dual-Head Knowledge Distillation: Enhancing Logits Utilization with an Auxiliary Head
Nov 13, 2024



Abstract:Traditional knowledge distillation focuses on aligning the student's predicted probabilities with both ground-truth labels and the teacher's predicted probabilities. However, the transition to predicted probabilities from logits would obscure certain indispensable information. To address this issue, it is intuitive to additionally introduce a logit-level loss function as a supplement to the widely used probability-level loss function, for exploiting the latent information of logits. Unfortunately, we empirically find that the amalgamation of the newly introduced logit-level loss and the previous probability-level loss will lead to performance degeneration, even trailing behind the performance of employing either loss in isolation. We attribute this phenomenon to the collapse of the classification head, which is verified by our theoretical analysis based on the neural collapse theory. Specifically, the gradients of the two loss functions exhibit contradictions in the linear classifier yet display no such conflict within the backbone. Drawing from the theoretical analysis, we propose a novel method called dual-head knowledge distillation, which partitions the linear classifier into two classification heads responsible for different losses, thereby preserving the beneficial effects of both losses on the backbone while eliminating adverse influences on the classification head. Extensive experiments validate that our method can effectively exploit the information inside the logits and achieve superior performance against state-of-the-art counterparts.
ChatCell: Facilitating Single-Cell Analysis with Natural Language
Feb 20, 2024



Abstract:As Large Language Models (LLMs) rapidly evolve, their influence in science is becoming increasingly prominent. The emerging capabilities of LLMs in task generalization and free-form dialogue can significantly advance fields like chemistry and biology. However, the field of single-cell biology, which forms the foundational building blocks of living organisms, still faces several challenges. High knowledge barriers and limited scalability in current methods restrict the full exploitation of LLMs in mastering single-cell data, impeding direct accessibility and rapid iteration. To this end, we introduce ChatCell, which signifies a paradigm shift by facilitating single-cell analysis with natural language. Leveraging vocabulary adaptation and unified sequence generation, ChatCell has acquired profound expertise in single-cell biology and the capability to accommodate a diverse range of analysis tasks. Extensive experiments further demonstrate ChatCell's robust performance and potential to deepen single-cell insights, paving the way for more accessible and intuitive exploration in this pivotal field. Our project homepage is available at https://zjunlp.github.io/project/ChatCell.
Multi-Label Knowledge Distillation
Aug 12, 2023
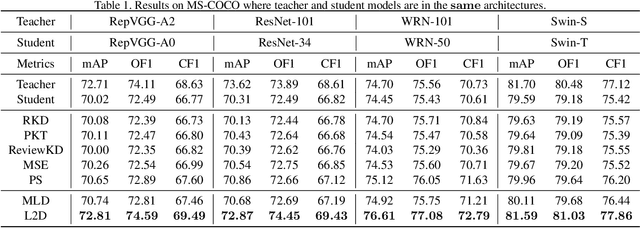
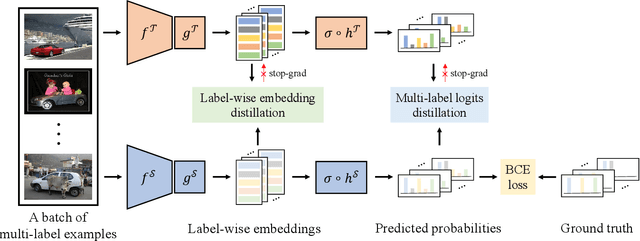
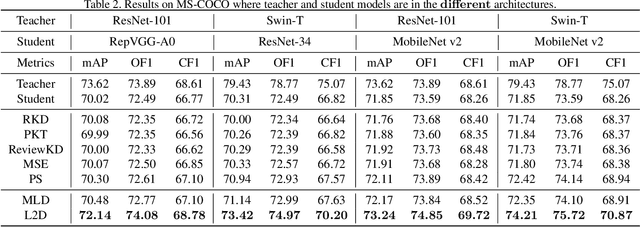
Abstract:Existing knowledge distillation methods typically work by imparting the knowledge of output logits or intermediate feature maps from the teacher network to the student network, which is very successful in multi-class single-label learning. However, these methods can hardly be extended to the multi-label learning scenario, where each instance is associated with multiple semantic labels, because the prediction probabilities do not sum to one and feature maps of the whole example may ignore minor classes in such a scenario. In this paper, we propose a novel multi-label knowledge distillation method. On one hand, it exploits the informative semantic knowledge from the logits by dividing the multi-label learning problem into a set of binary classification problems; on the other hand, it enhances the distinctiveness of the learned feature representations by leveraging the structural information of label-wise embeddings. Experimental results on multiple benchmark datasets validate that the proposed method can avoid knowledge counteraction among labels, thus achieving superior performance against diverse comparing methods. Our code is available at: https://github.com/penghui-yang/L2D
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge