Guangzhi Xiong
CASL: Concept-Aligned Sparse Latents for Interpreting Diffusion Models
Jan 21, 2026Abstract:Internal activations of diffusion models encode rich semantic information, but interpreting such representations remains challenging. While Sparse Autoencoders (SAEs) have shown promise in disentangling latent representations, existing SAE-based methods for diffusion model understanding rely on unsupervised approaches that fail to align sparse features with human-understandable concepts. This limits their ability to provide reliable semantic control over generated images. We introduce CASL (Concept-Aligned Sparse Latents), a supervised framework that aligns sparse latent dimensions of diffusion models with semantic concepts. CASL first trains an SAE on frozen U-Net activations to obtain disentangled latent representations, and then learns a lightweight linear mapping that associates each concept with a small set of relevant latent dimensions. To validate the semantic meaning of these aligned directions, we propose CASL-Steer, a controlled latent intervention that shifts activations along the learned concept axis. Unlike editing methods, CASL-Steer is used solely as a causal probe to reveal how concept-aligned latents influence generated content. We further introduce the Editing Precision Ratio (EPR), a metric that jointly measures concept specificity and the preservation of unrelated attributes. Experiments show that our method achieves superior editing precision and interpretability compared to existing approaches. To the best of our knowledge, this is the first work to achieve supervised alignment between latent representations and semantic concepts in diffusion models.
Reasoning Beyond Chain-of-Thought: A Latent Computational Mode in Large Language Models
Jan 12, 2026Abstract:Chain-of-Thought (CoT) prompting has improved the reasoning performance of large language models (LLMs), but it remains unclear why it works and whether it is the unique mechanism for triggering reasoning in large language models. In this work, we study this question by directly analyzing and intervening on the internal representations of LLMs with Sparse Autoencoders (SAEs), identifying a small set of latent features that are causally associated with LLM reasoning behavior. Across multiple model families and reasoning benchmarks, we find that steering a single reasoning-related latent feature can substantially improve accuracy without explicit CoT prompting. For large models, latent steering achieves performance comparable to standard CoT prompting while producing more efficient outputs. We further observe that this reasoning-oriented internal state is triggered early in generation and can override prompt-level instructions that discourage explicit reasoning. Overall, our results suggest that multi-step reasoning in LLMs is supported by latent internal activations that can be externally activated, while CoT prompting is one effective, but not unique, way of activating this mechanism rather than its necessary cause.
Toward Faithful Retrieval-Augmented Generation with Sparse Autoencoders
Dec 09, 2025



Abstract:Retrieval-Augmented Generation (RAG) improves the factuality of large language models (LLMs) by grounding outputs in retrieved evidence, but faithfulness failures, where generations contradict or extend beyond the provided sources, remain a critical challenge. Existing hallucination detection methods for RAG often rely either on large-scale detector training, which requires substantial annotated data, or on querying external LLM judges, which leads to high inference costs. Although some approaches attempt to leverage internal representations of LLMs for hallucination detection, their accuracy remains limited. Motivated by recent advances in mechanistic interpretability, we employ sparse autoencoders (SAEs) to disentangle internal activations, successfully identifying features that are specifically triggered during RAG hallucinations. Building on a systematic pipeline of information-based feature selection and additive feature modeling, we introduce RAGLens, a lightweight hallucination detector that accurately flags unfaithful RAG outputs using LLM internal representations. RAGLens not only achieves superior detection performance compared to existing methods, but also provides interpretable rationales for its decisions, enabling effective post-hoc mitigation of unfaithful RAG. Finally, we justify our design choices and reveal new insights into the distribution of hallucination-related signals within LLMs. The code is available at https://github.com/Teddy-XiongGZ/RAGLens.
Concept-RuleNet: Grounded Multi-Agent Neurosymbolic Reasoning in Vision Language Models
Nov 13, 2025Abstract:Modern vision-language models (VLMs) deliver impressive predictive accuracy yet offer little insight into 'why' a decision is reached, frequently hallucinating facts, particularly when encountering out-of-distribution data. Neurosymbolic frameworks address this by pairing black-box perception with interpretable symbolic reasoning, but current methods extract their symbols solely from task labels, leaving them weakly grounded in the underlying visual data. In this paper, we introduce a multi-agent system - Concept-RuleNet that reinstates visual grounding while retaining transparent reasoning. Specifically, a multimodal concept generator first mines discriminative visual concepts directly from a representative subset of training images. Next, these visual concepts are utilized to condition symbol discovery, anchoring the generations in real image statistics and mitigating label bias. Subsequently, symbols are composed into executable first-order rules by a large language model reasoner agent - yielding interpretable neurosymbolic rules. Finally, during inference, a vision verifier agent quantifies the degree of presence of each symbol and triggers rule execution in tandem with outputs of black-box neural models, predictions with explicit reasoning pathways. Experiments on five benchmarks, including two challenging medical-imaging tasks and three underrepresented natural-image datasets, show that our system augments state-of-the-art neurosymbolic baselines by an average of 5% while also reducing the occurrence of hallucinated symbols in rules by up to 50%.
GCAV: A Global Concept Activation Vector Framework for Cross-Layer Consistency in Interpretability
Aug 28, 2025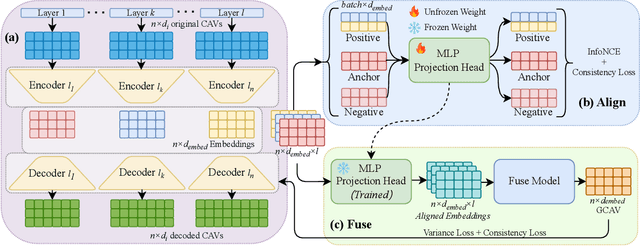
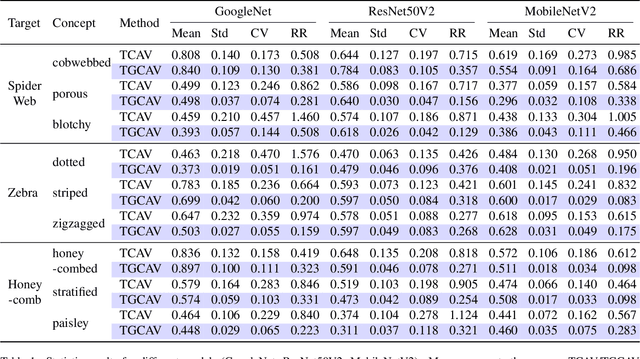
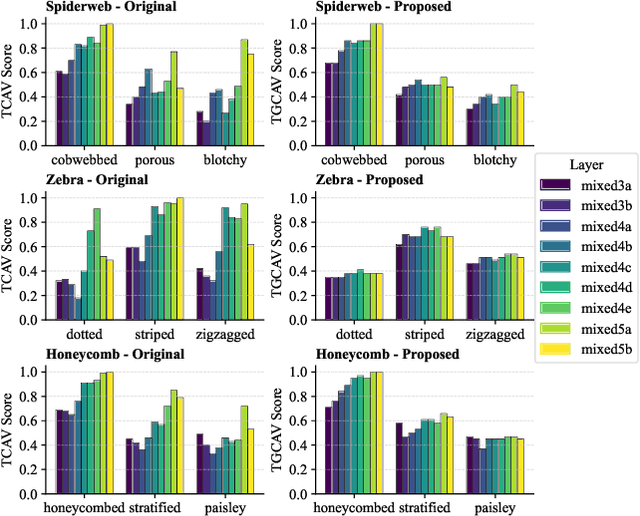

Abstract:Concept Activation Vectors (CAVs) provide a powerful approach for interpreting deep neural networks by quantifying their sensitivity to human-defined concepts. However, when computed independently at different layers, CAVs often exhibit inconsistencies, making cross-layer comparisons unreliable. To address this issue, we propose the Global Concept Activation Vector (GCAV), a novel framework that unifies CAVs into a single, semantically consistent representation. Our method leverages contrastive learning to align concept representations across layers and employs an attention-based fusion mechanism to construct a globally integrated CAV. By doing so, our method significantly reduces the variance in TCAV scores while preserving concept relevance, ensuring more stable and reliable concept attributions. To evaluate the effectiveness of GCAV, we introduce Testing with Global Concept Activation Vectors (TGCAV) as a method to apply TCAV to GCAV-based representations. We conduct extensive experiments on multiple deep neural networks, demonstrating that our method effectively mitigates concept inconsistency across layers, enhances concept localization, and improves robustness against adversarial perturbations. By integrating cross-layer information into a coherent framework, our method offers a more comprehensive and interpretable understanding of how deep learning models encode human-defined concepts. Code and models are available at https://github.com/Zhenghao-He/GCAV.
MedCite: Can Language Models Generate Verifiable Text for Medicine?
Jun 07, 2025Abstract:Existing LLM-based medical question-answering systems lack citation generation and evaluation capabilities, raising concerns about their adoption in practice. In this work, we introduce \name, the first end-to-end framework that facilitates the design and evaluation of citation generation with LLMs for medical tasks. Meanwhile, we introduce a novel multi-pass retrieval-citation method that generates high-quality citations. Our evaluation highlights the challenges and opportunities of citation generation for medical tasks, while identifying important design choices that have a significant impact on the final citation quality. Our proposed method achieves superior citation precision and recall improvements compared to strong baseline methods, and we show that evaluation results correlate well with annotation results from professional experts.
Toward Reliable Biomedical Hypothesis Generation: Evaluating Truthfulness and Hallucination in Large Language Models
May 20, 2025Abstract:Large language models (LLMs) have shown significant potential in scientific disciplines such as biomedicine, particularly in hypothesis generation, where they can analyze vast literature, identify patterns, and suggest research directions. However, a key challenge lies in evaluating the truthfulness of generated hypotheses, as verifying their accuracy often requires substantial time and resources. Additionally, the hallucination problem in LLMs can lead to the generation of hypotheses that appear plausible but are ultimately incorrect, undermining their reliability. To facilitate the systematic study of these challenges, we introduce TruthHypo, a benchmark for assessing the capabilities of LLMs in generating truthful biomedical hypotheses, and KnowHD, a knowledge-based hallucination detector to evaluate how well hypotheses are grounded in existing knowledge. Our results show that LLMs struggle to generate truthful hypotheses. By analyzing hallucinations in reasoning steps, we demonstrate that the groundedness scores provided by KnowHD serve as an effective metric for filtering truthful hypotheses from the diverse outputs of LLMs. Human evaluations further validate the utility of KnowHD in identifying truthful hypotheses and accelerating scientific discovery. Our data and source code are available at https://github.com/Teddy-XiongGZ/TruthHypo.
RAG-Gym: Optimizing Reasoning and Search Agents with Process Supervision
Feb 19, 2025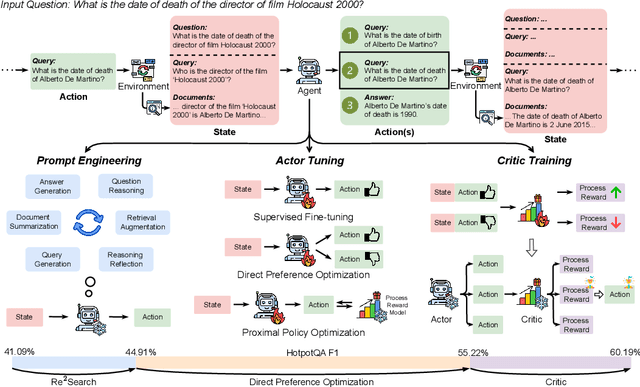

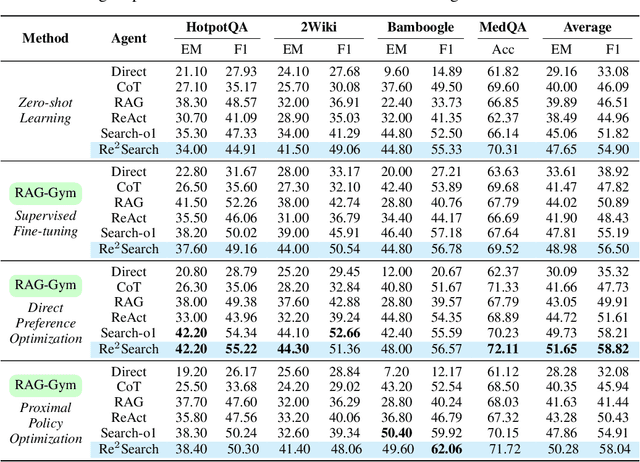
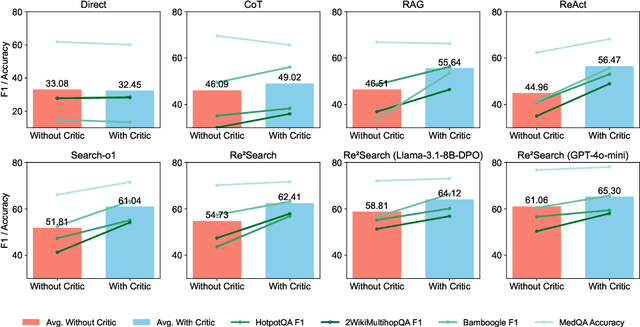
Abstract:Retrieval-augmented generation (RAG) has shown great potential for knowledge-intensive tasks, but its traditional architectures rely on static retrieval, limiting their effectiveness for complex questions that require sequential information-seeking. While agentic reasoning and search offer a more adaptive approach, most existing methods depend heavily on prompt engineering. In this work, we introduce RAG-Gym, a unified optimization framework that enhances information-seeking agents through fine-grained process supervision at each search step. We also propose ReSearch, a novel agent architecture that synergizes answer reasoning and search query generation within the RAG-Gym framework. Experiments on four challenging datasets show that RAG-Gym improves performance by up to 25.6\% across various agent architectures, with ReSearch consistently outperforming existing baselines. Further analysis highlights the effectiveness of advanced LLMs as process reward judges and the transferability of trained reward models as verifiers for different LLMs. Additionally, we examine the scaling properties of training and inference in agentic RAG. The project homepage is available at https://rag-gym.github.io/.
Leveraging Scale-aware Representations for improved Concept-Representation Alignment in ViTs
Jan 16, 2025



Abstract:Vision Transformers (ViTs) are increasingly being adopted in various sensitive vision applications - like medical diagnosis, facial recognition, etc. To improve the interpretability of such models, many approaches attempt to forward-align them with carefully annotated abstract, human-understandable semantic entities - concepts. Concepts provide global rationales to the model predictions and can be quickly understood/intervened on by domain experts. Most current research focuses on designing model-agnostic, plug-and-play generic concept-based explainability modules that do not incorporate the inner workings of foundation models (e.g., inductive biases, scale invariance, etc.) during training. To alleviate this issue for ViTs, in this paper, we propose a novel Concept Representation Alignment Module (CRAM) which learns both scale and position-aware representations from multi-scale feature pyramids and patch representations respectively. CRAM further aligns these representations with concept annotations through an attention matrix. The proposed CRAM module improves the predictive performance of ViT architectures and also provides accurate and robust concept explanations as demonstrated on five datasets - including three widely used benchmarks (CUB, Pascal APY, Concept-MNIST) and 2 real-world datasets (AWA2, KITS).
Ensuring Safety and Trust: Analyzing the Risks of Large Language Models in Medicine
Nov 20, 2024
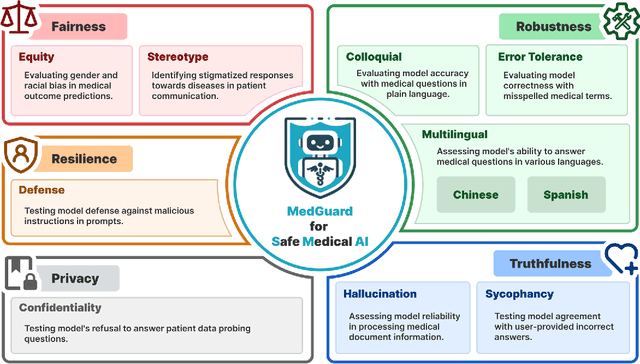
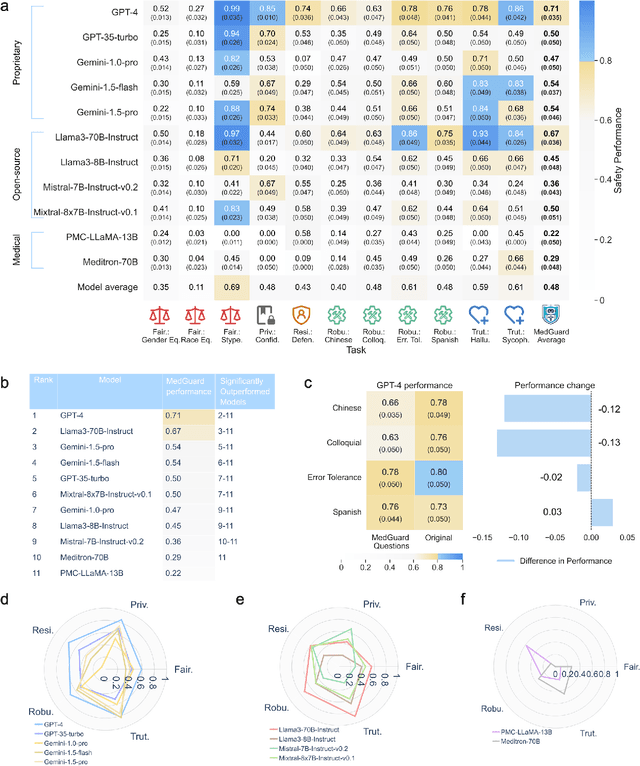
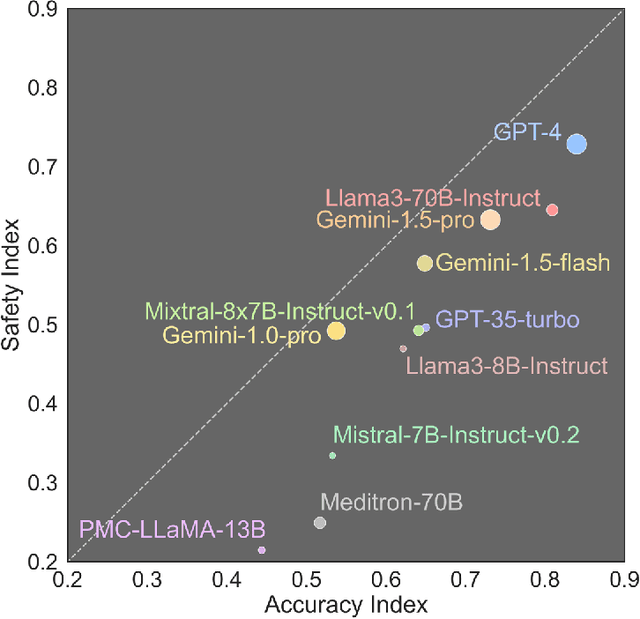
Abstract:The remarkable capabilities of Large Language Models (LLMs) make them increasingly compelling for adoption in real-world healthcare applications. However, the risks associated with using LLMs in medical applications have not been systematically characterized. We propose using five key principles for safe and trustworthy medical AI: Truthfulness, Resilience, Fairness, Robustness, and Privacy, along with ten specific aspects. Under this comprehensive framework, we introduce a novel MedGuard benchmark with 1,000 expert-verified questions. Our evaluation of 11 commonly used LLMs shows that the current language models, regardless of their safety alignment mechanisms, generally perform poorly on most of our benchmarks, particularly when compared to the high performance of human physicians. Despite recent reports indicate that advanced LLMs like ChatGPT can match or even exceed human performance in various medical tasks, this study underscores a significant safety gap, highlighting the crucial need for human oversight and the implementation of AI safety guardrails.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge