W. John Wilbur
Ensuring Safety and Trust: Analyzing the Risks of Large Language Models in Medicine
Nov 20, 2024
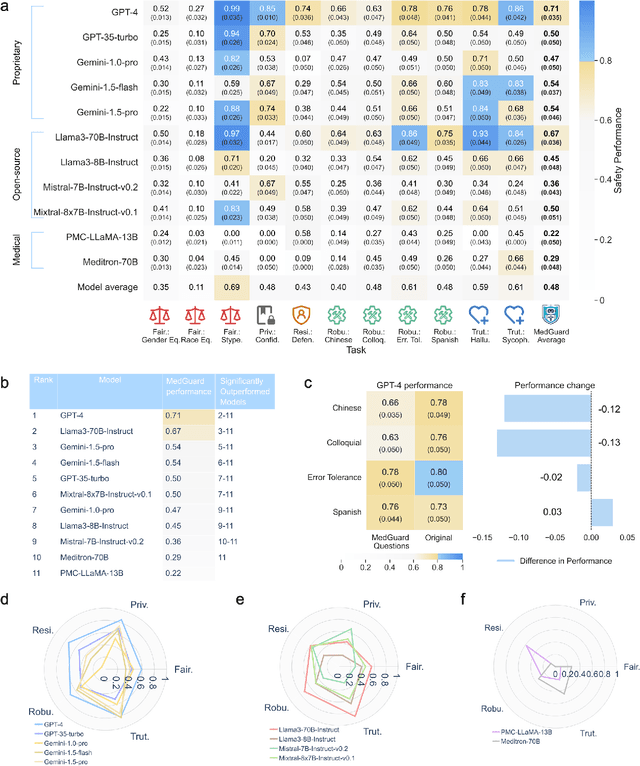
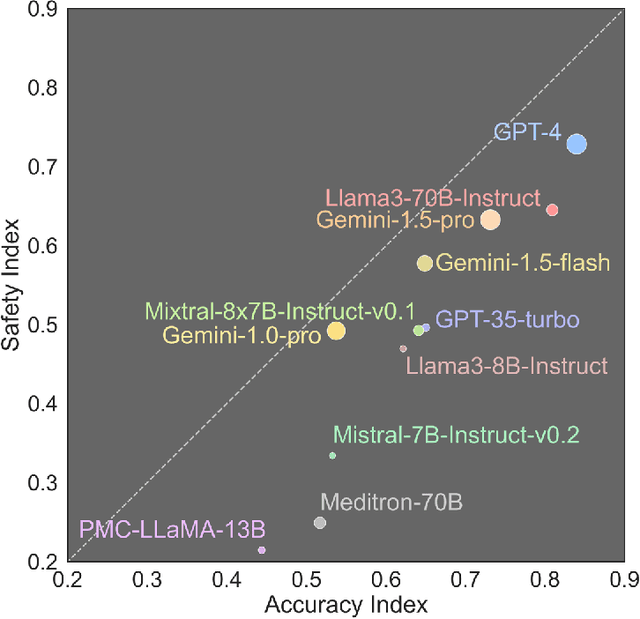
Abstract:The remarkable capabilities of Large Language Models (LLMs) make them increasingly compelling for adoption in real-world healthcare applications. However, the risks associated with using LLMs in medical applications have not been systematically characterized. We propose using five key principles for safe and trustworthy medical AI: Truthfulness, Resilience, Fairness, Robustness, and Privacy, along with ten specific aspects. Under this comprehensive framework, we introduce a novel MedGuard benchmark with 1,000 expert-verified questions. Our evaluation of 11 commonly used LLMs shows that the current language models, regardless of their safety alignment mechanisms, generally perform poorly on most of our benchmarks, particularly when compared to the high performance of human physicians. Despite recent reports indicate that advanced LLMs like ChatGPT can match or even exceed human performance in various medical tasks, this study underscores a significant safety gap, highlighting the crucial need for human oversight and the implementation of AI safety guardrails.
Comprehensive identification of Long Covid articles with human-in-the-loop machine learning
Sep 16, 2022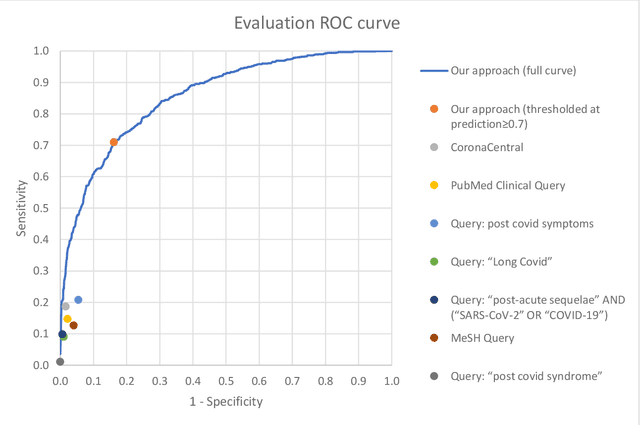
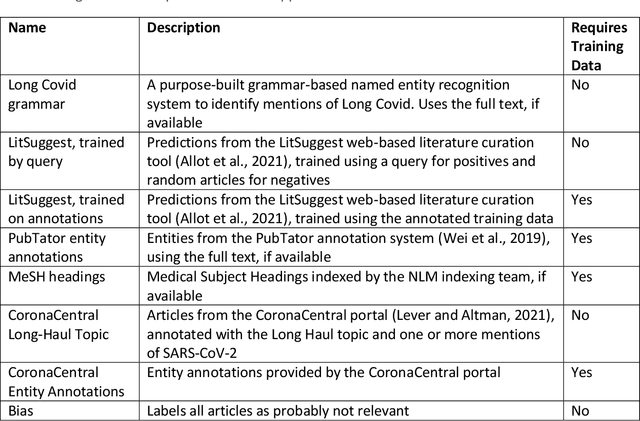
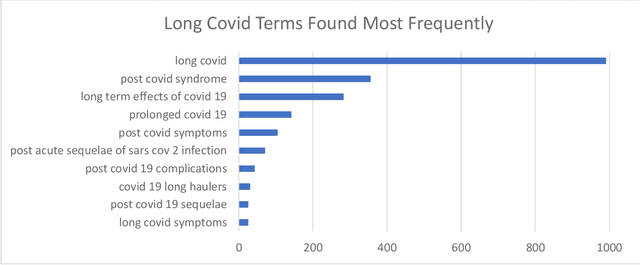
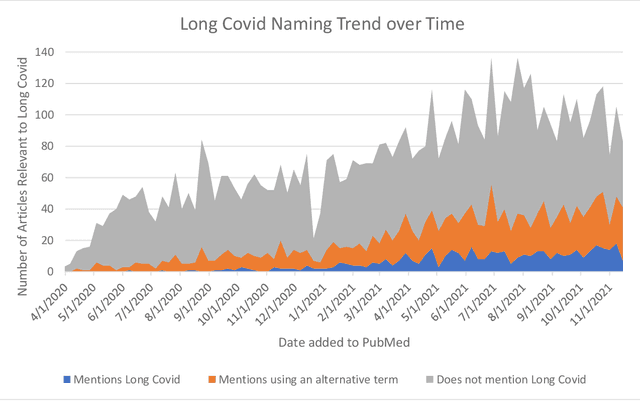
Abstract:A significant percentage of COVID-19 survivors experience ongoing multisystemic symptoms that often affect daily living, a condition known as Long Covid or post-acute-sequelae of SARS-CoV-2 infection. However, identifying Long Covid articles is challenging since articles refer to the condition using a variety of less common terms or refrain from naming it at all. We developed an iterative human-in-the-loop machine learning framework designed to effectively leverage the data available and make the most efficient use of human labels. Specifically, our approach combines data programming with active learning into a robust ensemble model. Evaluating our model on a holdout set demonstrates over three times the sensitivity of other methods. We apply our model to PubMed to create the Long Covid collection, and demonstrate that (1) most Long Covid articles do not refer to Long Covid by any name (2) when the condition is named, the name used most frequently in the biomedical literature is Long Covid, and (3) Long Covid is associated with disorders in a wide variety of body systems. The Long Covid collection is updated weekly and is searchable online at the LitCovid portal: https://www.ncbi.nlm.nih.gov/research/coronavirus/docsum?filters=e_condition.LongCovid
Navigating the landscape of COVID-19 research through literature analysis: A bird's eye view
Sep 11, 2020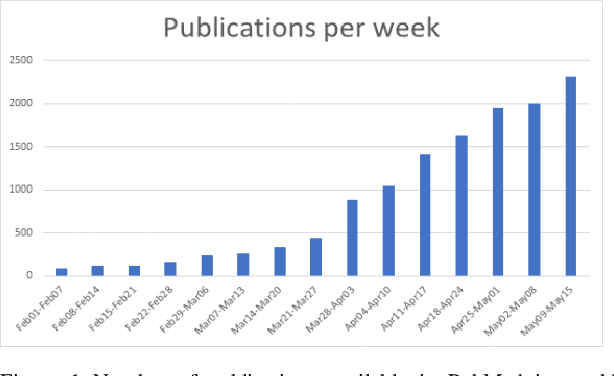
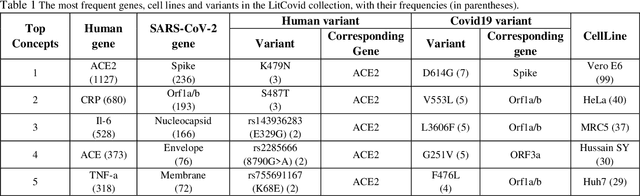
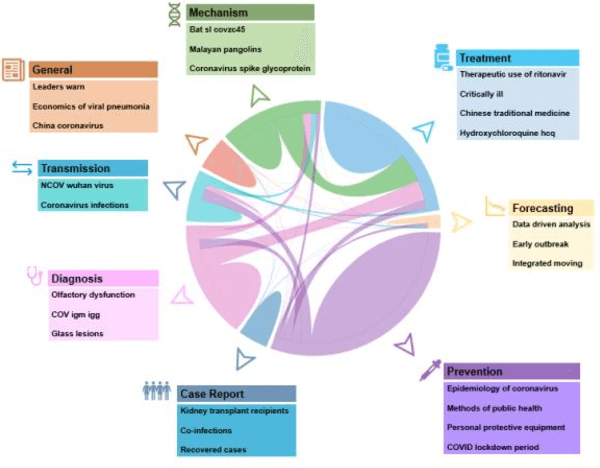
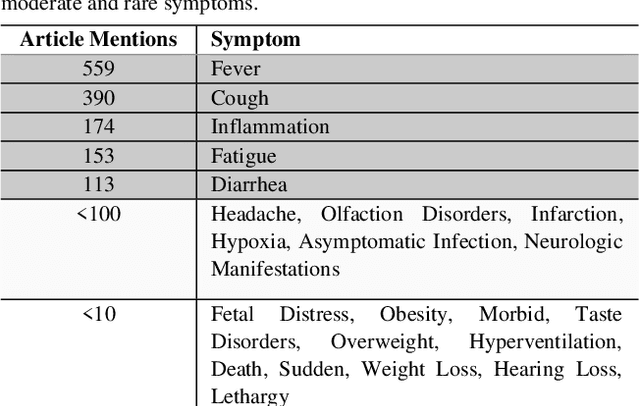
Abstract:Timely access to accurate scientific literature in the battle with the ongoing COVID-19 pandemic is critical. This unprecedented public health risk has motivated research towards understanding the disease in general, identifying drugs to treat the disease, developing potential vaccines, etc. This has given rise to a rapidly growing body of literature that doubles in number of publications every 20 days as of May 2020. Providing medical professionals with means to quickly analyze the literature and discover growing areas of knowledge is necessary for addressing their question and information needs. In this study we analyze the LitCovid collection, 13,369 COVID-19 related articles found in PubMed as of May 15th, 2020 with the purpose of examining the landscape of literature and presenting it in a format that facilitates information navigation and understanding. We do that by applying state-of-the-art named entity recognition, classification, clustering and other NLP techniques. By applying NER tools, we capture relevant bioentities (such as diseases, internal body organs, etc.) and assess the strength of their relationship with COVID-19 by the extent they are discussed in the corpus. We also collect a variety of symptoms and co-morbidities discussed in reference to COVID-19. Our clustering algorithm identifies topics represented by groups of related terms, and computes clusters corresponding to documents associated with the topic terms. Among the topics we observe several that persist through the duration of multiple weeks and have numerous associated documents, as well several that appear as emerging topics with fewer documents. All the tools and data are publicly available, and this framework can be applied to any literature collection. Taken together, these analyses produce a comprehensive, synthesized view of COVID-19 research to facilitate knowledge discovery from literature.
* 10 pages, 8 Figures, Submitted to KDD 2020 Health Day
PDC -- a probabilistic distributional clustering algorithm: a case study on suicide articles in PubMed
Dec 04, 2019
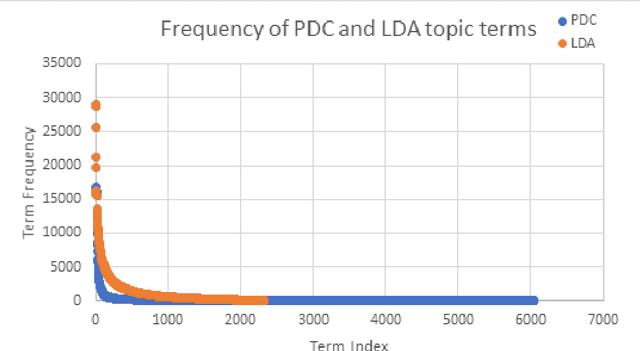
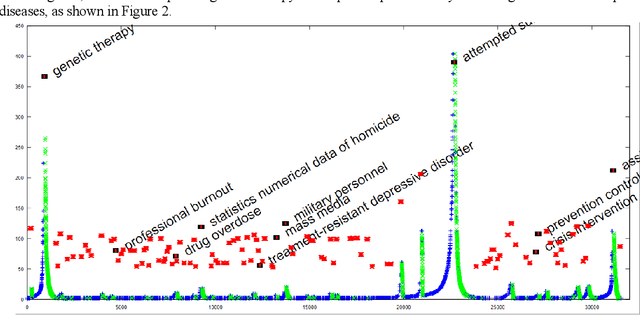

Abstract:The need to organize a large collection in a manner that facilitates human comprehension is crucial given the ever-increasing volumes of information. In this work, we present PDC (probabilistic distributional clustering), a novel algorithm that, given a document collection, computes disjoint term sets representing topics in the collection. The algorithm relies on probabilities of word co-occurrences to partition the set of terms appearing in the collection of documents into disjoint groups of related terms. In this work, we also present an environment to visualize the computed topics in the term space and retrieve the most related PubMed articles for each group of terms. We illustrate the algorithm by applying it to PubMed documents on the topic of suicide. Suicide is a major public health problem identified as the tenth leading cause of death in the US. In this application, our goal is to provide a global view of the mental health literature pertaining to the subject of suicide, and through this, to help create a rich environment of multifaceted data to guide health care researchers in their endeavor to better understand the breadth, depth and scope of the problem. We demonstrate the usefulness of the proposed algorithm by providing a web portal that allows mental health researchers to peruse the suicide-related literature in PubMed.
Deep learning with sentence embeddings pre-trained on biomedical corpora improves the performance of finding similar sentences in electronic medical records
Sep 06, 2019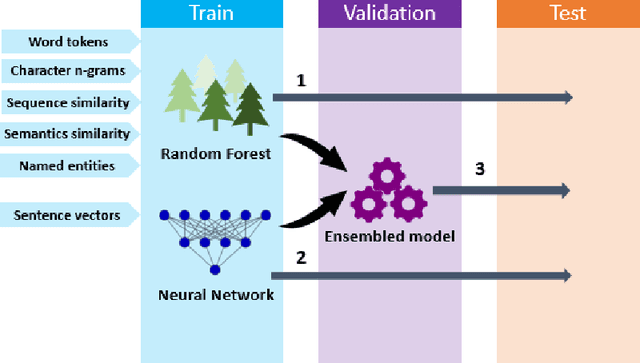
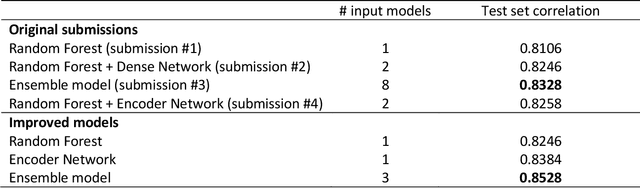
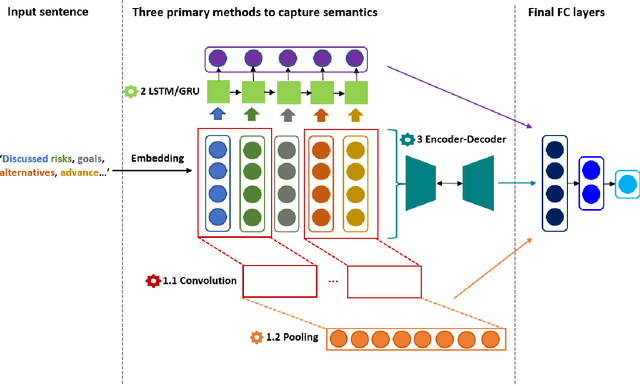
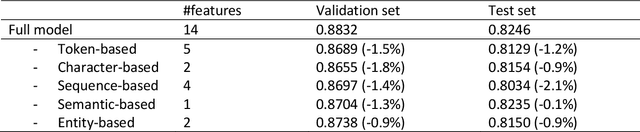
Abstract:Capturing sentence semantics plays a vital role in a range of text mining applications. Despite continuous efforts on the development of related datasets and models in the general domain, both datasets and models are limited in biomedical and clinical domains. The BioCreative/OHNLP organizers have made the first attempt to annotate 1,068 sentence pairs from clinical notes and have called for a community effort to tackle the Semantic Textual Similarity (BioCreative/OHNLP STS) challenge. We developed models using traditional machine learning and deep learning approaches. For the post challenge, we focus on two models: the Random Forest and the Encoder Network. We applied sentence embeddings pre-trained on PubMed abstracts and MIMIC-III clinical notes and updated the Random Forest and the Encoder Network accordingly. The official results demonstrated our best submission was the ensemble of eight models. It achieved a Person correlation coefficient of 0.8328, the highest performance among 13 submissions from 4 teams. For the post challenge, the performance of both Random Forest and the Encoder Network was improved; in particular, the correlation of the Encoder Network was improved by ~13%. During the challenge task, no end-to-end deep learning models had better performance than machine learning models that take manually-crafted features. In contrast, with the sentence embeddings pre-trained on biomedical corpora, the Encoder Network now achieves a correlation of ~0.84, which is higher than the original best model. The ensembled model taking the improved versions of the Random Forest and Encoder Network as inputs further increased performance to 0.8528. Deep learning models with sentence embeddings pre-trained on biomedical corpora achieve the highest performance on the test set.
Bridging the Gap: Incorporating a Semantic Similarity Measure for Effectively Mapping PubMed Queries to Documents
Oct 17, 2017
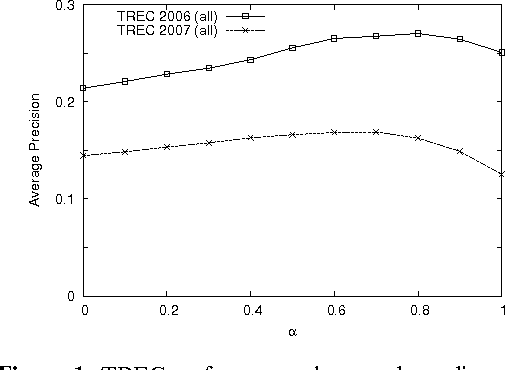

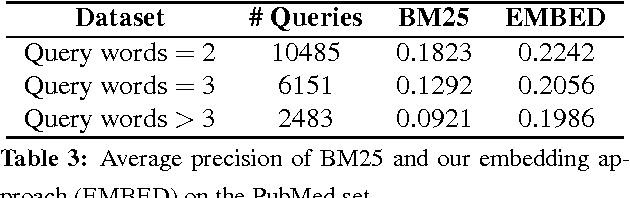
Abstract:The main approach of traditional information retrieval (IR) is to examine how many words from a query appear in a document. A drawback of this approach, however, is that it may fail to detect relevant documents where no or only few words from a query are found. The semantic analysis methods such as LSA (latent semantic analysis) and LDA (latent Dirichlet allocation) have been proposed to address the issue, but their performance is not superior compared to common IR approaches. Here we present a query-document similarity measure motivated by the Word Mover's Distance. Unlike other similarity measures, the proposed method relies on neural word embeddings to compute the distance between words. This process helps identify related words when no direct matches are found between a query and a document. Our method is efficient and straightforward to implement. The experimental results on TREC Genomics data show that our approach outperforms the BM25 ranking function by an average of 12% in mean average precision. Furthermore, for a real-world dataset collected from the PubMed search logs, we combine the semantic measure with BM25 using a learning to rank method, which leads to improved ranking scores by up to 25%. This experiment demonstrates that the proposed approach and BM25 nicely complement each other and together produce superior performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge