Lana Yeganova
BioCPT: Contrastive Pre-trained Transformers with Large-scale PubMed Search Logs for Zero-shot Biomedical Information Retrieval
Jul 02, 2023Abstract:Information retrieval (IR) is essential in biomedical knowledge acquisition and clinical decision support. While recent progress has shown that language model encoders perform better semantic retrieval, training such models requires abundant query-article annotations that are difficult to obtain in biomedicine. As a result, most biomedical IR systems only conduct lexical matching. In response, we introduce BioCPT, a first-of-its-kind Contrastively Pre-trained Transformer model for zero-shot biomedical IR. To train BioCPT, we collected an unprecedented scale of 255 million user click logs from PubMed. With such data, we use contrastive learning to train a pair of closely-integrated retriever and re-ranker. Experimental results show that BioCPT sets new state-of-the-art performance on five biomedical IR tasks, outperforming various baselines including much larger models such as GPT-3-sized cpt-text-XL. In addition, BioCPT also generates better biomedical article and sentence representations for semantic evaluations. As such, BioCPT can be readily applied to various real-world biomedical IR tasks. BioCPT API and code are publicly available at https://github.com/ncbi/BioCPT.
Opportunities and Challenges for ChatGPT and Large Language Models in Biomedicine and Health
Jun 15, 2023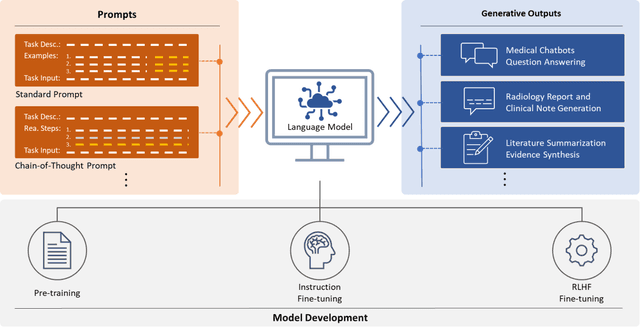



Abstract:ChatGPT has drawn considerable attention from both the general public and domain experts with its remarkable text generation capabilities. This has subsequently led to the emergence of diverse applications in the field of biomedicine and health. In this work, we examine the diverse applications of large language models (LLMs), such as ChatGPT, in biomedicine and health. Specifically we explore the areas of biomedical information retrieval, question answering, medical text summarization, information extraction, and medical education, and investigate whether LLMs possess the transformative power to revolutionize these tasks or whether the distinct complexities of biomedical domain presents unique challenges. Following an extensive literature survey, we find that significant advances have been made in the field of text generation tasks, surpassing the previous state-of-the-art methods. For other applications, the advances have been modest. Overall, LLMs have not yet revolutionized the biomedicine, but recent rapid progress indicates that such methods hold great potential to provide valuable means for accelerating discovery and improving health. We also find that the use of LLMs, like ChatGPT, in the fields of biomedicine and health entails various risks and challenges, including fabricated information in its generated responses, as well as legal and privacy concerns associated with sensitive patient data. We believe this first-of-its-kind survey can provide a comprehensive overview to biomedical researchers and healthcare practitioners on the opportunities and challenges associated with using ChatGPT and other LLMs for transforming biomedicine and health.
Navigating the landscape of COVID-19 research through literature analysis: A bird's eye view
Sep 11, 2020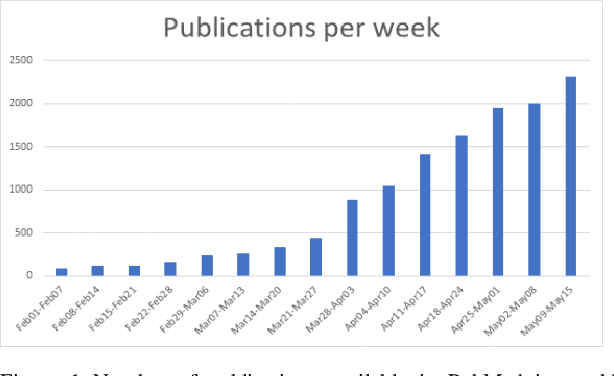
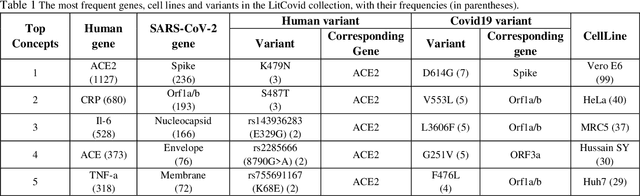
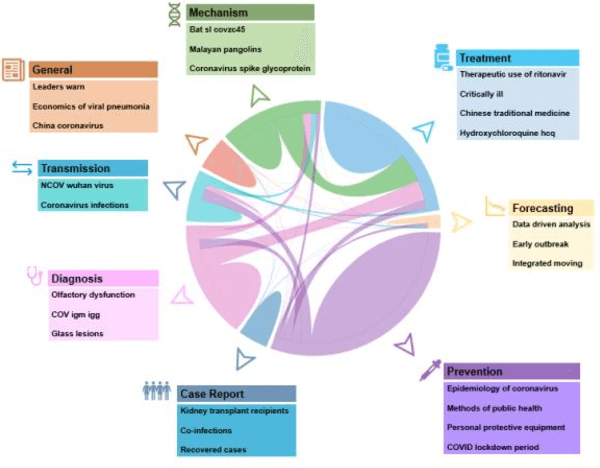
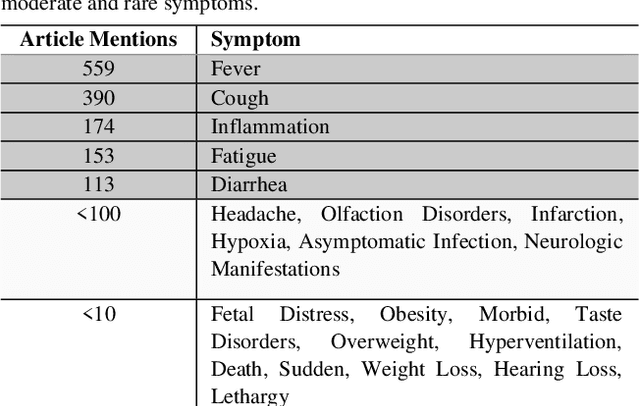
Abstract:Timely access to accurate scientific literature in the battle with the ongoing COVID-19 pandemic is critical. This unprecedented public health risk has motivated research towards understanding the disease in general, identifying drugs to treat the disease, developing potential vaccines, etc. This has given rise to a rapidly growing body of literature that doubles in number of publications every 20 days as of May 2020. Providing medical professionals with means to quickly analyze the literature and discover growing areas of knowledge is necessary for addressing their question and information needs. In this study we analyze the LitCovid collection, 13,369 COVID-19 related articles found in PubMed as of May 15th, 2020 with the purpose of examining the landscape of literature and presenting it in a format that facilitates information navigation and understanding. We do that by applying state-of-the-art named entity recognition, classification, clustering and other NLP techniques. By applying NER tools, we capture relevant bioentities (such as diseases, internal body organs, etc.) and assess the strength of their relationship with COVID-19 by the extent they are discussed in the corpus. We also collect a variety of symptoms and co-morbidities discussed in reference to COVID-19. Our clustering algorithm identifies topics represented by groups of related terms, and computes clusters corresponding to documents associated with the topic terms. Among the topics we observe several that persist through the duration of multiple weeks and have numerous associated documents, as well several that appear as emerging topics with fewer documents. All the tools and data are publicly available, and this framework can be applied to any literature collection. Taken together, these analyses produce a comprehensive, synthesized view of COVID-19 research to facilitate knowledge discovery from literature.
* 10 pages, 8 Figures, Submitted to KDD 2020 Health Day
PDC -- a probabilistic distributional clustering algorithm: a case study on suicide articles in PubMed
Dec 04, 2019
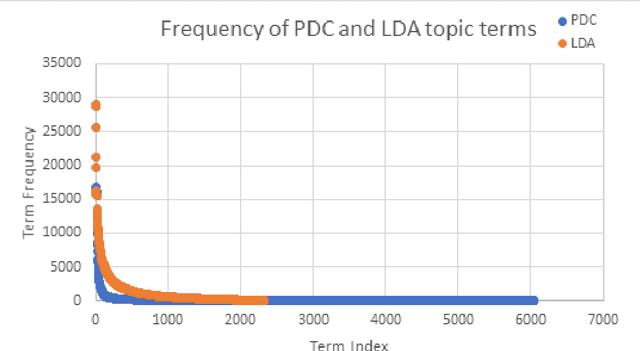
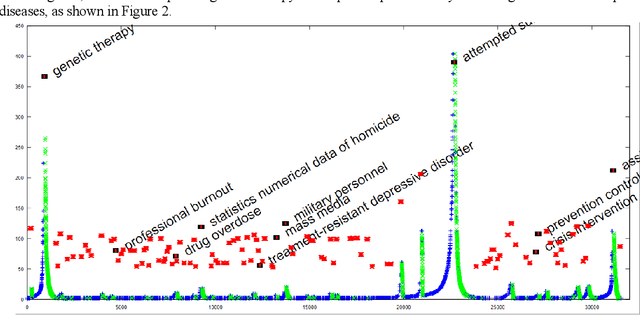

Abstract:The need to organize a large collection in a manner that facilitates human comprehension is crucial given the ever-increasing volumes of information. In this work, we present PDC (probabilistic distributional clustering), a novel algorithm that, given a document collection, computes disjoint term sets representing topics in the collection. The algorithm relies on probabilities of word co-occurrences to partition the set of terms appearing in the collection of documents into disjoint groups of related terms. In this work, we also present an environment to visualize the computed topics in the term space and retrieve the most related PubMed articles for each group of terms. We illustrate the algorithm by applying it to PubMed documents on the topic of suicide. Suicide is a major public health problem identified as the tenth leading cause of death in the US. In this application, our goal is to provide a global view of the mental health literature pertaining to the subject of suicide, and through this, to help create a rich environment of multifaceted data to guide health care researchers in their endeavor to better understand the breadth, depth and scope of the problem. We demonstrate the usefulness of the proposed algorithm by providing a web portal that allows mental health researchers to peruse the suicide-related literature in PubMed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge