Shubo Tian
Knowledge-guided Contextual Gene Set Analysis Using Large Language Models
Jun 04, 2025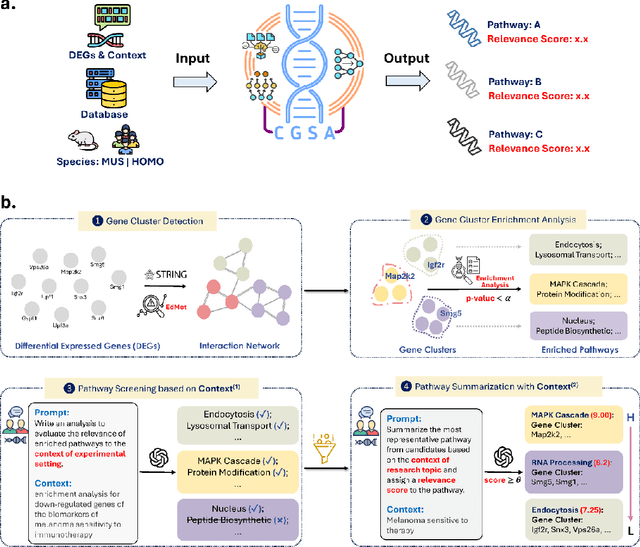

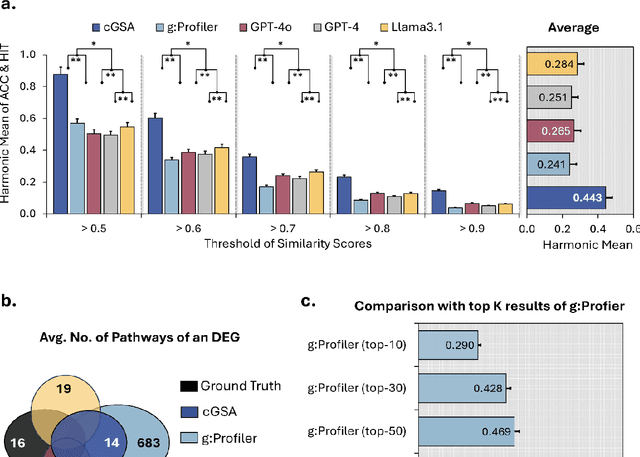

Abstract:Gene set analysis (GSA) is a foundational approach for interpreting genomic data of diseases by linking genes to biological processes. However, conventional GSA methods overlook clinical context of the analyses, often generating long lists of enriched pathways with redundant, nonspecific, or irrelevant results. Interpreting these requires extensive, ad-hoc manual effort, reducing both reliability and reproducibility. To address this limitation, we introduce cGSA, a novel AI-driven framework that enhances GSA by incorporating context-aware pathway prioritization. cGSA integrates gene cluster detection, enrichment analysis, and large language models to identify pathways that are not only statistically significant but also biologically meaningful. Benchmarking on 102 manually curated gene sets across 19 diseases and ten disease-related biological mechanisms shows that cGSA outperforms baseline methods by over 30%, with expert validation confirming its increased precision and interpretability. Two independent case studies in melanoma and breast cancer further demonstrate its potential to uncover context-specific insights and support targeted hypothesis generation.
Enhancing Biomedical Relation Extraction with Directionality
Jan 23, 2025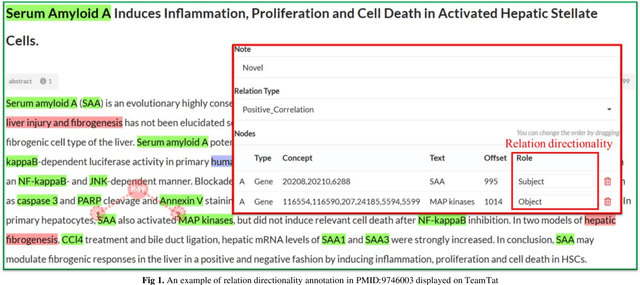
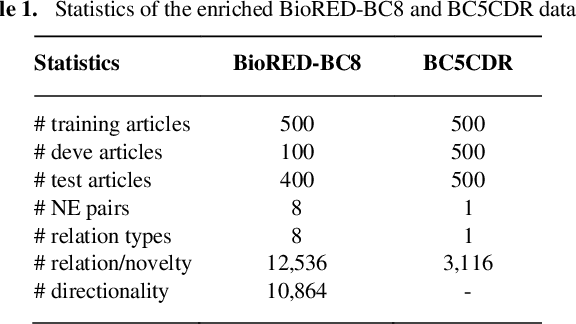
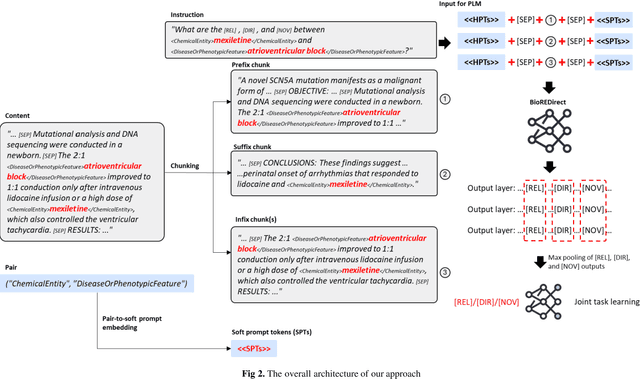
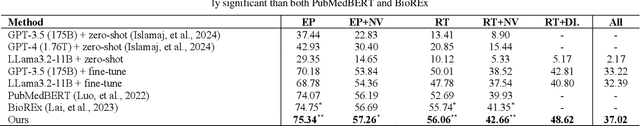
Abstract:Biological relation networks contain rich information for understanding the biological mechanisms behind the relationship of entities such as genes, proteins, diseases, and chemicals. The vast growth of biomedical literature poses significant challenges updating the network knowledge. The recent Biomedical Relation Extraction Dataset (BioRED) provides valuable manual annotations, facilitating the develop-ment of machine-learning and pre-trained language model approaches for automatically identifying novel document-level (inter-sentence context) relationships. Nonetheless, its annotations lack directionality (subject/object) for the entity roles, essential for studying complex biological networks. Herein we annotate the entity roles of the relationships in the BioRED corpus and subsequently propose a novel multi-task language model with soft-prompt learning to jointly identify the relationship, novel findings, and entity roles. Our results in-clude an enriched BioRED corpus with 10,864 directionality annotations. Moreover, our proposed method outperforms existing large language models such as the state-of-the-art GPT-4 and Llama-3 on two benchmarking tasks. Our source code and dataset are available at https://github.com/ncbi-nlp/BioREDirect.
Demystifying Large Language Models for Medicine: A Primer
Oct 24, 2024



Abstract:Large language models (LLMs) represent a transformative class of AI tools capable of revolutionizing various aspects of healthcare by generating human-like responses across diverse contexts and adapting to novel tasks following human instructions. Their potential application spans a broad range of medical tasks, such as clinical documentation, matching patients to clinical trials, and answering medical questions. In this primer paper, we propose an actionable guideline to help healthcare professionals more efficiently utilize LLMs in their work, along with a set of best practices. This approach consists of several main phases, including formulating the task, choosing LLMs, prompt engineering, fine-tuning, and deployment. We start with the discussion of critical considerations in identifying healthcare tasks that align with the core capabilities of LLMs and selecting models based on the selected task and data, performance requirements, and model interface. We then review the strategies, such as prompt engineering and fine-tuning, to adapt standard LLMs to specialized medical tasks. Deployment considerations, including regulatory compliance, ethical guidelines, and continuous monitoring for fairness and bias, are also discussed. By providing a structured step-by-step methodology, this tutorial aims to equip healthcare professionals with the tools necessary to effectively integrate LLMs into clinical practice, ensuring that these powerful technologies are applied in a safe, reliable, and impactful manner.
GeneAgent: Self-verification Language Agent for Gene Set Knowledge Discovery using Domain Databases
May 25, 2024



Abstract:Gene set knowledge discovery is essential for advancing human functional genomics. Recent studies have shown promising performance by harnessing the power of Large Language Models (LLMs) on this task. Nonetheless, their results are subject to several limitations common in LLMs such as hallucinations. In response, we present GeneAgent, a first-of-its-kind language agent featuring self-verification capability. It autonomously interacts with various biological databases and leverages relevant domain knowledge to improve accuracy and reduce hallucination occurrences. Benchmarking on 1,106 gene sets from different sources, GeneAgent consistently outperforms standard GPT-4 by a significant margin. Moreover, a detailed manual review confirms the effectiveness of the self-verification module in minimizing hallucinations and generating more reliable analytical narratives. To demonstrate its practical utility, we apply GeneAgent to seven novel gene sets derived from mouse B2905 melanoma cell lines, with expert evaluations showing that GeneAgent offers novel insights into gene functions and subsequently expedites knowledge discovery.
Quality of Answers of Generative Large Language Models vs Peer Patients for Interpreting Lab Test Results for Lay Patients: Evaluation Study
Jan 23, 2024Abstract:Lab results are often confusing and hard to understand. Large language models (LLMs) such as ChatGPT have opened a promising avenue for patients to get their questions answered. We aim to assess the feasibility of using LLMs to generate relevant, accurate, helpful, and unharmful responses to lab test-related questions asked by patients and to identify potential issues that can be mitigated with augmentation approaches. We first collected lab test results related question and answer data from Yahoo! Answers and selected 53 QA pairs for this study. Using the LangChain framework and ChatGPT web portal, we generated responses to the 53 questions from four LLMs including GPT-4, Meta LLaMA 2, MedAlpaca, and ORCA_mini. We first assessed the similarity of their answers using standard QA similarity-based evaluation metrics including ROUGE, BLEU, METEOR, BERTScore. We also utilized an LLM-based evaluator to judge whether a target model has higher quality in terms of relevance, correctness, helpfulness, and safety than the baseline model. Finally, we performed a manual evaluation with medical experts for all the responses to seven selected questions on the same four aspects. The results of Win Rate and medical expert evaluation both showed that GPT-4's responses achieved better scores than all the other LLM responses and human responses on all four aspects (relevance, correctness, helpfulness, and safety). However, LLM responses occasionally also suffer from a lack of interpretation in one's medical context, incorrect statements, and lack of references. We find that compared to other three LLMs and human answer from the Q&A website, GPT-4's responses are more accurate, helpful, relevant, and safer. However, there are cases which GPT-4 responses are inaccurate and not individualized. We identified a number of ways to improve the quality of LLM responses.
PubTator 3.0: an AI-powered Literature Resource for Unlocking Biomedical Knowledge
Jan 19, 2024

Abstract:PubTator 3.0 (https://www.ncbi.nlm.nih.gov/research/pubtator3/) is a biomedical literature resource using state-of-the-art AI techniques to offer semantic and relation searches for key concepts like proteins, genetic variants, diseases, and chemicals. It currently provides over one billion entity and relation annotations across approximately 36 million PubMed abstracts and 6 million full-text articles from the PMC open access subset, updated weekly. PubTator 3.0's online interface and API utilize these precomputed entity relations and synonyms to provide advanced search capabilities and enable large-scale analyses, streamlining many complex information needs. We showcase the retrieval quality of PubTator 3.0 using a series of entity pair queries, demonstrating that PubTator 3.0 retrieves a greater number of articles than either PubMed or Google Scholar, with higher precision in the top 20 results. We further show that integrating ChatGPT (GPT-4) with PubTator APIs dramatically improves the factuality and verifiability of its responses. In summary, PubTator 3.0 offers a comprehensive set of features and tools that allow researchers to navigate the ever-expanding wealth of biomedical literature, expediting research and unlocking valuable insights for scientific discovery.
Information Retrieval and Classification of Real-Time Multi-Source Hurricane Evacuation Notices
Jan 07, 2024Abstract:For an approaching disaster, the tracking of time-sensitive critical information such as hurricane evacuation notices is challenging in the United States. These notices are issued and distributed rapidly by numerous local authorities that may spread across multiple states. They often undergo frequent updates and are distributed through diverse online portals lacking standard formats. In this study, we developed an approach to timely detect and track the locally issued hurricane evacuation notices. The text data were collected mainly with a spatially targeted web scraping method. They were manually labeled and then classified using natural language processing techniques with deep learning models. The classification of mandatory evacuation notices achieved a high accuracy (recall = 96%). We used Hurricane Ian (2022) to illustrate how real-time evacuation notices extracted from local government sources could be redistributed with a Web GIS system. Our method applied to future hurricanes provides live data for situation awareness to higher-level government agencies and news media. The archived data helps scholars to study government responses toward weather warnings and individual behaviors influenced by evacuation history. The framework may be applied to other types of disasters for rapid and targeted retrieval, classification, redistribution, and archiving of real-time government orders and notifications.
Opportunities and Challenges for ChatGPT and Large Language Models in Biomedicine and Health
Jun 15, 2023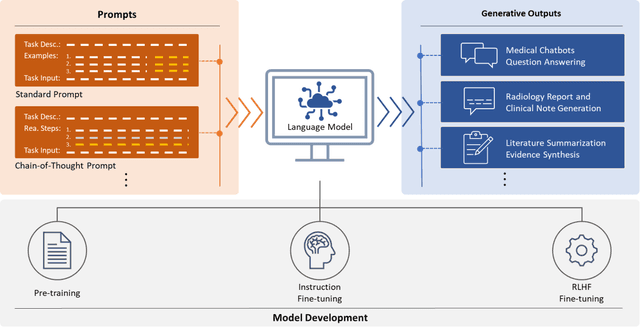



Abstract:ChatGPT has drawn considerable attention from both the general public and domain experts with its remarkable text generation capabilities. This has subsequently led to the emergence of diverse applications in the field of biomedicine and health. In this work, we examine the diverse applications of large language models (LLMs), such as ChatGPT, in biomedicine and health. Specifically we explore the areas of biomedical information retrieval, question answering, medical text summarization, information extraction, and medical education, and investigate whether LLMs possess the transformative power to revolutionize these tasks or whether the distinct complexities of biomedical domain presents unique challenges. Following an extensive literature survey, we find that significant advances have been made in the field of text generation tasks, surpassing the previous state-of-the-art methods. For other applications, the advances have been modest. Overall, LLMs have not yet revolutionized the biomedicine, but recent rapid progress indicates that such methods hold great potential to provide valuable means for accelerating discovery and improving health. We also find that the use of LLMs, like ChatGPT, in the fields of biomedicine and health entails various risks and challenges, including fabricated information in its generated responses, as well as legal and privacy concerns associated with sensitive patient data. We believe this first-of-its-kind survey can provide a comprehensive overview to biomedical researchers and healthcare practitioners on the opportunities and challenges associated with using ChatGPT and other LLMs for transforming biomedicine and health.
Multi-label classification for biomedical literature: an overview of the BioCreative VII LitCovid Track for COVID-19 literature topic annotations
Apr 20, 2022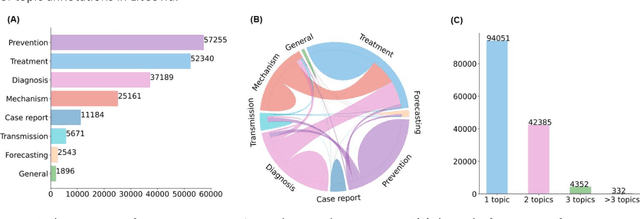
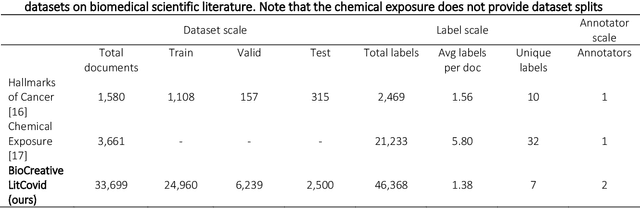
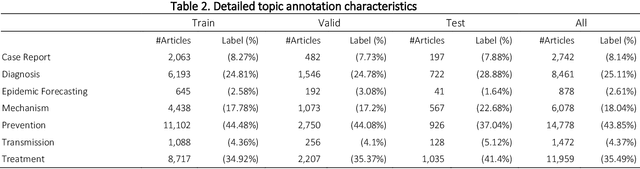
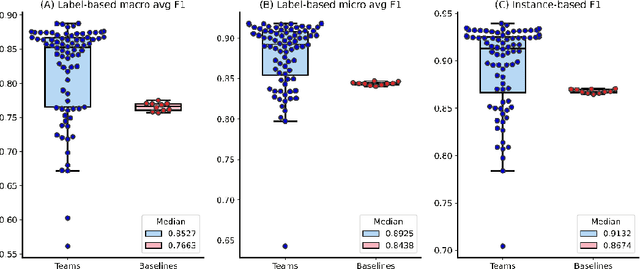
Abstract:The COVID-19 pandemic has been severely impacting global society since December 2019. Massive research has been undertaken to understand the characteristics of the virus and design vaccines and drugs. The related findings have been reported in biomedical literature at a rate of about 10,000 articles on COVID-19 per month. Such rapid growth significantly challenges manual curation and interpretation. For instance, LitCovid is a literature database of COVID-19-related articles in PubMed, which has accumulated more than 200,000 articles with millions of accesses each month by users worldwide. One primary curation task is to assign up to eight topics (e.g., Diagnosis and Treatment) to the articles in LitCovid. Despite the continuing advances in biomedical text mining methods, few have been dedicated to topic annotations in COVID-19 literature. To close the gap, we organized the BioCreative LitCovid track to call for a community effort to tackle automated topic annotation for COVID-19 literature. The BioCreative LitCovid dataset, consisting of over 30,000 articles with manually reviewed topics, was created for training and testing. It is one of the largest multilabel classification datasets in biomedical scientific literature. 19 teams worldwide participated and made 80 submissions in total. Most teams used hybrid systems based on transformers. The highest performing submissions achieved 0.8875, 0.9181, and 0.9394 for macro F1-score, micro F1-score, and instance-based F1-score, respectively. The level of participation and results demonstrate a successful track and help close the gap between dataset curation and method development. The dataset is publicly available via https://ftp.ncbi.nlm.nih.gov/pub/lu/LitCovid/biocreative/ for benchmarking and further development.
Temporal Subtyping of Alzheimer's Disease Using Medical Conditions Preceding Alzheimer's Disease Onset in Electronic Health Records
Feb 22, 2022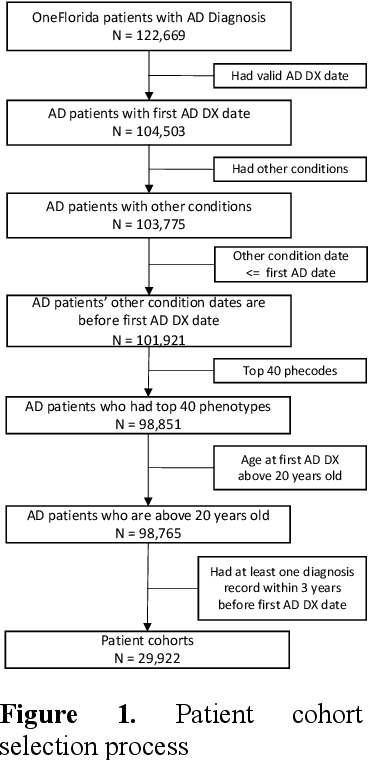
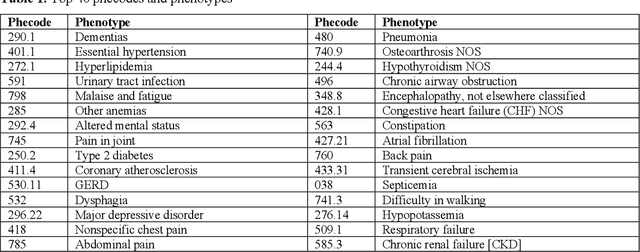
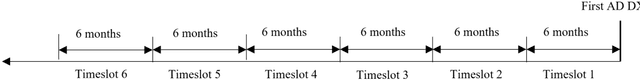
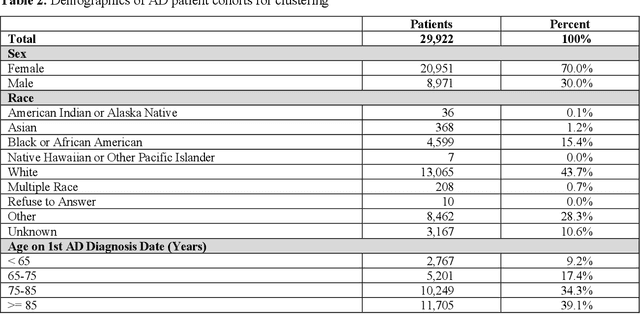
Abstract:Subtyping of Alzheimer's disease (AD) can facilitate diagnosis, treatment, prognosis and disease management. It can also support the testing of new prevention and treatment strategies through clinical trials. In this study, we employed spectral clustering to cluster 29,922 AD patients in the OneFlorida Data Trust using their longitudinal EHR data of diagnosis and conditions into four subtypes. These subtypes exhibit different patterns of progression of other conditions prior to the first AD diagnosis. In addition, according to the results of various statistical tests, these subtypes are also significantly different with respect to demographics, mortality, and prescription medications after the AD diagnosis. This study could potentially facilitate early detection and personalized treatment of AD as well as data-driven generalizability assessment of clinical trials for AD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge