Ling Luo
AFD-INSTRUCTION: A Comprehensive Antibody Instruction Dataset with Functional Annotations for LLM-Based Understanding and Design
Feb 04, 2026Abstract:Large language models (LLMs) have significantly advanced protein representation learning. However, their capacity to interpret and design antibodies through natural language remains limited. To address this challenge, we present AFD-Instruction, the first large-scale instruction dataset with functional annotations tailored to antibodies. This dataset encompasses two key components: antibody understanding, which infers functional attributes directly from sequences, and antibody design, which enables de novo sequence generation under functional constraints. These components provide explicit sequence-function alignment and support antibody design guided by natural language instructions. Extensive instruction-tuning experiments on general-purpose LLMs demonstrate that AFD-Instruction consistently improves performance across diverse antibody-related tasks. By linking antibody sequences with textual descriptions of function, AFD-Instruction establishes a new foundation for advancing antibody modeling and accelerating therapeutic discovery.
Stability as a Liability:Systematic Breakdown of Linguistic Structure in LLMs
Jan 26, 2026Abstract:Training stability is typically regarded as a prerequisite for reliable optimization in large language models. In this work, we analyze how stabilizing training dynamics affects the induced generation distribution. We show that under standard maximum likelihood training, stable parameter trajectories lead stationary solutions to approximately minimize the forward KL divergence to the empirical distribution, while implicitly reducing generative entropy. As a consequence, the learned model can concentrate probability mass on a limited subset of empirical modes, exhibiting systematic degeneration despite smooth loss convergence. We empirically validate this effect using a controlled feedback-based training framework that stabilizes internal generation statistics, observing consistent low-entropy outputs and repetitive behavior across architectures and random seeds. It indicates that optimization stability and generative expressivity are not inherently aligned, and that stability alone is an insufficient indicator of generative quality.
Overview of CHIP 2025 Shared Task 2: Discharge Medication Recommendation for Metabolic Diseases Based on Chinese Electronic Health Records
Nov 09, 2025Abstract:Discharge medication recommendation plays a critical role in ensuring treatment continuity, preventing readmission, and improving long-term management for patients with chronic metabolic diseases. This paper present an overview of the CHIP 2025 Shared Task 2 competition, which aimed to develop state-of-the-art approaches for automatically recommending appro-priate discharge medications using real-world Chinese EHR data. For this task, we constructed CDrugRed, a high-quality dataset consisting of 5,894 de-identified hospitalization records from 3,190 patients in China. This task is challenging due to multi-label nature of medication recommendation, het-erogeneous clinical text, and patient-specific variability in treatment plans. A total of 526 teams registered, with 167 and 95 teams submitting valid results to the Phase A and Phase B leaderboards, respectively. The top-performing team achieved the highest overall performance on the final test set, with a Jaccard score of 0.5102, F1 score of 0.6267, demonstrating the potential of advanced large language model (LLM)-based ensemble systems. These re-sults highlight both the promise and remaining challenges of applying LLMs to medication recommendation in Chinese EHRs. The post-evaluation phase remains open at https://tianchi.aliyun.com/competition/entrance/532411/.
MedTrust-RAG: Evidence Verification and Trust Alignment for Biomedical Question Answering
Oct 16, 2025Abstract:Biomedical question answering (QA) requires accurate interpretation of complex medical knowledge. Large language models (LLMs) have shown promising capabilities in this domain, with retrieval-augmented generation (RAG) systems enhancing performance by incorporating external medical literature. However, RAG-based approaches in biomedical QA suffer from hallucinations due to post-retrieval noise and insufficient verification of retrieved evidence, undermining response reliability. We propose MedTrust-Guided Iterative RAG, a framework designed to enhance factual consistency and mitigate hallucinations in medical QA. Our method introduces three key innovations. First, it enforces citation-aware reasoning by requiring all generated content to be explicitly grounded in retrieved medical documents, with structured Negative Knowledge Assertions used when evidence is insufficient. Second, it employs an iterative retrieval-verification process, where a verification agent assesses evidence adequacy and refines queries through Medical Gap Analysis until reliable information is obtained. Third, it integrates the MedTrust-Align Module (MTAM) that combines verified positive examples with hallucination-aware negative samples, leveraging Direct Preference Optimization to reinforce citation-grounded reasoning while penalizing hallucination-prone response patterns. Experiments on MedMCQA, MedQA, and MMLU-Med demonstrate that our approach consistently outperforms competitive baselines across multiple model architectures, achieving the best average accuracy with gains of 2.7% for LLaMA3.1-8B-Instruct and 2.4% for Qwen3-8B.
FocusMed: A Large Language Model-based Framework for Enhancing Medical Question Summarization with Focus Identification
Oct 06, 2025
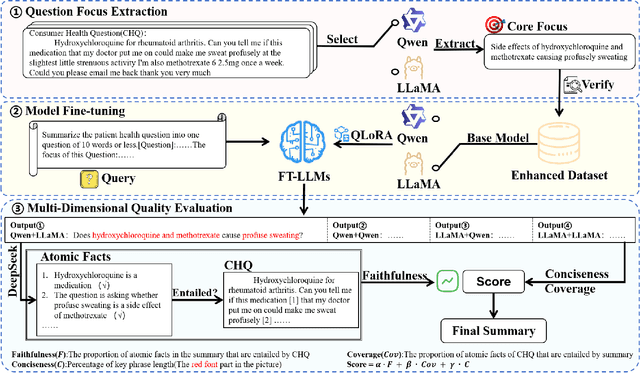

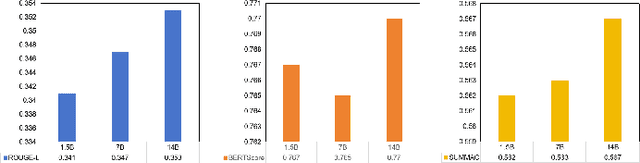
Abstract:With the rapid development of online medical platforms, consumer health questions (CHQs) are inefficient in diagnosis due to redundant information and frequent non-professional terms. The medical question summary (MQS) task aims to transform CHQs into streamlined doctors' frequently asked questions (FAQs), but existing methods still face challenges such as poor identification of question focus and model hallucination. This paper explores the potential of large language models (LLMs) in the MQS task and finds that direct fine-tuning is prone to focus identification bias and generates unfaithful content. To this end, we propose an optimization framework based on core focus guidance. First, a prompt template is designed to drive the LLMs to extract the core focus from the CHQs that is faithful to the original text. Then, a fine-tuning dataset is constructed in combination with the original CHQ-FAQ pairs to improve the ability to identify the focus of the question. Finally, a multi-dimensional quality evaluation and selection mechanism is proposed to comprehensively improve the quality of the summary from multiple dimensions. We conduct comprehensive experiments on two widely-adopted MQS datasets using three established evaluation metrics. The proposed framework achieves state-of-the-art performance across all measures, demonstrating a significant boost in the model's ability to identify critical focus of questions and a notable mitigation of hallucinations. The source codes are freely available at https://github.com/DUT-LiuChao/FocusMed.
ViewCraft3D: High-Fidelity and View-Consistent 3D Vector Graphics Synthesis
May 26, 2025Abstract:3D vector graphics play a crucial role in various applications including 3D shape retrieval, conceptual design, and virtual reality interactions due to their ability to capture essential structural information with minimal representation. While recent approaches have shown promise in generating 3D vector graphics, they often suffer from lengthy processing times and struggle to maintain view consistency. To address these limitations, we propose ViewCraft3D (VC3D), an efficient method that leverages 3D priors to generate 3D vector graphics. Specifically, our approach begins with 3D object analysis, employs a geometric extraction algorithm to fit 3D vector graphics to the underlying structure, and applies view-consistent refinement process to enhance visual quality. Our comprehensive experiments demonstrate that VC3D outperforms previous methods in both qualitative and quantitative evaluations, while significantly reducing computational overhead. The resulting 3D sketches maintain view consistency and effectively capture the essential characteristics of the original objects.
Dynamical Label Augmentation and Calibration for Noisy Electronic Health Records
May 12, 2025Abstract:Medical research, particularly in predicting patient outcomes, heavily relies on medical time series data extracted from Electronic Health Records (EHR), which provide extensive information on patient histories. Despite rigorous examination, labeling errors are inevitable and can significantly impede accurate predictions of patient outcome. To address this challenge, we propose an \textbf{A}ttention-based Learning Framework with Dynamic \textbf{C}alibration and Augmentation for \textbf{T}ime series Noisy \textbf{L}abel \textbf{L}earning (ACTLL). This framework leverages a two-component Beta mixture model to identify the certain and uncertain sets of instances based on the fitness distribution of each class, and it captures global temporal dynamics while dynamically calibrating labels from the uncertain set or augmenting confident instances from the certain set. Experimental results on large-scale EHR datasets eICU and MIMIC-IV-ED, and several benchmark datasets from the UCR and UEA repositories, demonstrate that our model ACTLL has achieved state-of-the-art performance, especially under high noise levels.
SHIP: A Shapelet-based Approach for Interpretable Patient-Ventilator Asynchrony Detection
Mar 09, 2025Abstract:Patient-ventilator asynchrony (PVA) is a common and critical issue during mechanical ventilation, affecting up to 85% of patients. PVA can result in clinical complications such as discomfort, sleep disruption, and potentially more severe conditions like ventilator-induced lung injury and diaphragm dysfunction. Traditional PVA management, which relies on manual adjustments by healthcare providers, is often inadequate due to delays and errors. While various computational methods, including rule-based, statistical, and deep learning approaches, have been developed to detect PVA events, they face challenges related to dataset imbalances and lack of interpretability. In this work, we propose a shapelet-based approach SHIP for PVA detection, utilizing shapelets - discriminative subsequences in time-series data - to enhance detection accuracy and interpretability. Our method addresses dataset imbalances through shapelet-based data augmentation and constructs a shapelet pool to transform the dataset for more effective classification. The combined shapelet and statistical features are then used in a classifier to identify PVA events. Experimental results on medical datasets show that SHIP significantly improves PVA detection while providing interpretable insights into model decisions.
Large Language Model Driven Agents for Simulating Echo Chamber Formation
Feb 25, 2025Abstract:The rise of echo chambers on social media platforms has heightened concerns about polarization and the reinforcement of existing beliefs. Traditional approaches for simulating echo chamber formation have often relied on predefined rules and numerical simulations, which, while insightful, may lack the nuance needed to capture complex, real-world interactions. In this paper, we present a novel framework that leverages large language models (LLMs) as generative agents to simulate echo chamber dynamics within social networks. The novelty of our approach is that it incorporates both opinion updates and network rewiring behaviors driven by LLMs, allowing for a context-aware and semantically rich simulation of social interactions. Additionally, we utilize real-world Twitter (now X) data to benchmark the LLM-based simulation against actual social media behaviors, providing insights into the accuracy and realism of the generated opinion trends. Our results demonstrate the efficacy of LLMs in modeling echo chamber formation, capturing both structural and semantic dimensions of opinion clustering. %This work contributes to a deeper understanding of social influence dynamics and offers a new tool for studying polarization in online communities.
SVGS-DSGAT: An IoT-Enabled Innovation in Underwater Robotic Object Detection Technology
Jan 21, 2025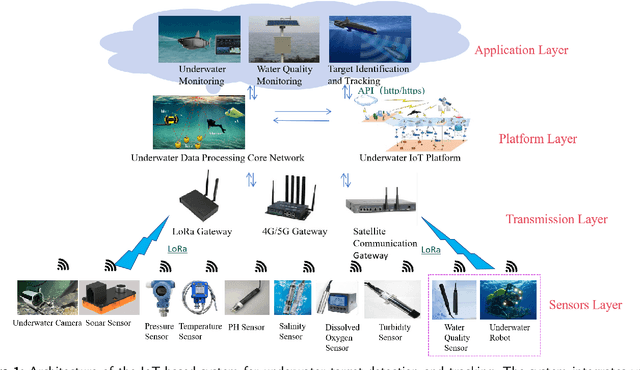
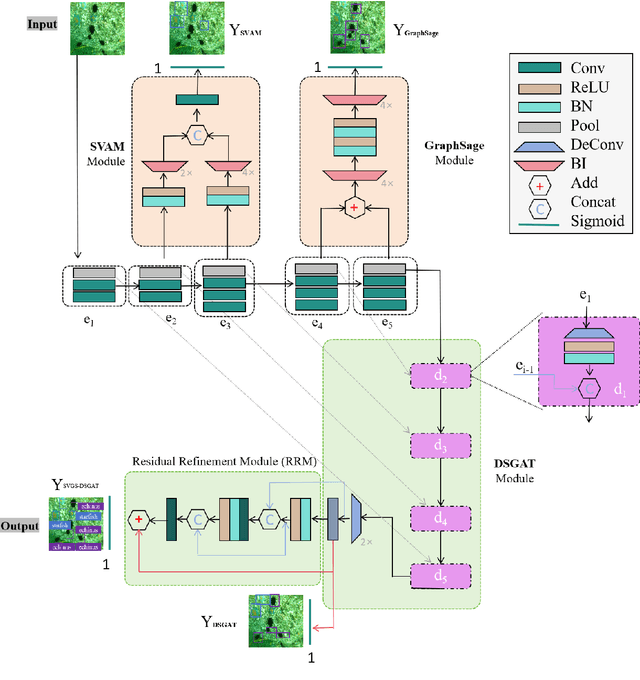
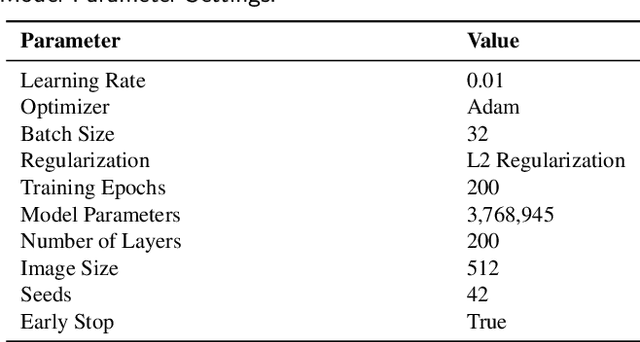
Abstract:With the advancement of Internet of Things (IoT) technology, underwater target detection and tracking have become increasingly important for ocean monitoring and resource management. Existing methods often fall short in handling high-noise and low-contrast images in complex underwater environments, lacking precision and robustness. This paper introduces a novel SVGS-DSGAT model that combines GraphSage, SVAM, and DSGAT modules, enhancing feature extraction and target detection capabilities through graph neural networks and attention mechanisms. The model integrates IoT technology to facilitate real-time data collection and processing, optimizing resource allocation and model responsiveness. Experimental results demonstrate that the SVGS-DSGAT model achieves an mAP of 40.8% on the URPC 2020 dataset and 41.5% on the SeaDronesSee dataset, significantly outperforming existing mainstream models. This IoT-enhanced approach not only excels in high-noise and complex backgrounds but also improves the overall efficiency and scalability of the system. This research provides an effective IoT solution for underwater target detection technology, offering significant practical application value and broad development prospects.
* 17 pages, 8 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge