Shuo Xu
TACOcc:Target-Adaptive Cross-Modal Fusion with Volume Rendering for 3D Semantic Occupancy
May 19, 2025Abstract:The performance of multi-modal 3D occupancy prediction is limited by ineffective fusion, mainly due to geometry-semantics mismatch from fixed fusion strategies and surface detail loss caused by sparse, noisy annotations. The mismatch stems from the heterogeneous scale and distribution of point cloud and image features, leading to biased matching under fixed neighborhood fusion. To address this, we propose a target-scale adaptive, bidirectional symmetric retrieval mechanism. It expands the neighborhood for large targets to enhance context awareness and shrinks it for small ones to improve efficiency and suppress noise, enabling accurate cross-modal feature alignment. This mechanism explicitly establishes spatial correspondences and improves fusion accuracy. For surface detail loss, sparse labels provide limited supervision, resulting in poor predictions for small objects. We introduce an improved volume rendering pipeline based on 3D Gaussian Splatting, which takes fused features as input to render images, applies photometric consistency supervision, and jointly optimizes 2D-3D consistency. This enhances surface detail reconstruction while suppressing noise propagation. In summary, we propose TACOcc, an adaptive multi-modal fusion framework for 3D semantic occupancy prediction, enhanced by volume rendering supervision. Experiments on the nuScenes and SemanticKITTI benchmarks validate its effectiveness.
LLM-based Automated Theorem Proving Hinges on Scalable Synthetic Data Generation
May 17, 2025Abstract:Recent advancements in large language models (LLMs) have sparked considerable interest in automated theorem proving and a prominent line of research integrates stepwise LLM-based provers into tree search. In this paper, we introduce a novel proof-state exploration approach for training data synthesis, designed to produce diverse tactics across a wide range of intermediate proof states, thereby facilitating effective one-shot fine-tuning of LLM as the policy model. We also propose an adaptive beam size strategy, which effectively takes advantage of our data synthesis method and achieves a trade-off between exploration and exploitation during tree search. Evaluations on the MiniF2F and ProofNet benchmarks demonstrate that our method outperforms strong baselines under the stringent Pass@1 metric, attaining an average pass rate of $60.74\%$ on MiniF2F and $21.18\%$ on ProofNet. These results underscore the impact of large-scale synthetic data in advancing automated theorem proving.
Correlative and Discriminative Label Grouping for Multi-Label Visual Prompt Tuning
Apr 14, 2025Abstract:Modeling label correlations has always played a pivotal role in multi-label image classification (MLC), attracting significant attention from researchers. However, recent studies have overemphasized co-occurrence relationships among labels, which can lead to overfitting risk on this overemphasis, resulting in suboptimal models. To tackle this problem, we advocate for balancing correlative and discriminative relationships among labels to mitigate the risk of overfitting and enhance model performance. To this end, we propose the Multi-Label Visual Prompt Tuning framework, a novel and parameter-efficient method that groups classes into multiple class subsets according to label co-occurrence and mutual exclusivity relationships, and then models them respectively to balance the two relationships. In this work, since each group contains multiple classes, multiple prompt tokens are adopted within Vision Transformer (ViT) to capture the correlation or discriminative label relationship within each group, and effectively learn correlation or discriminative representations for class subsets. On the other hand, each group contains multiple group-aware visual representations that may correspond to multiple classes, and the mixture of experts (MoE) model can cleverly assign them from the group-aware to the label-aware, adaptively obtaining label-aware representation, which is more conducive to classification. Experiments on multiple benchmark datasets show that our proposed approach achieves competitive results and outperforms SOTA methods on multiple pre-trained models.
Headache to Overstock? Promoting Long-tail Items through Debiased Product Bundling
Nov 28, 2024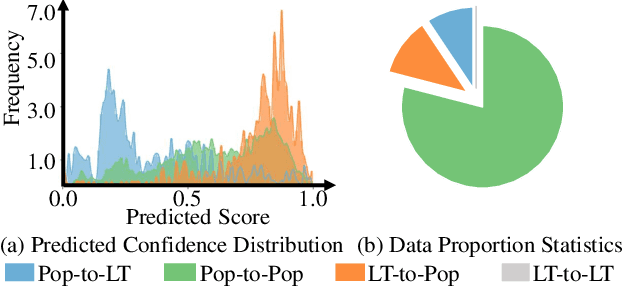

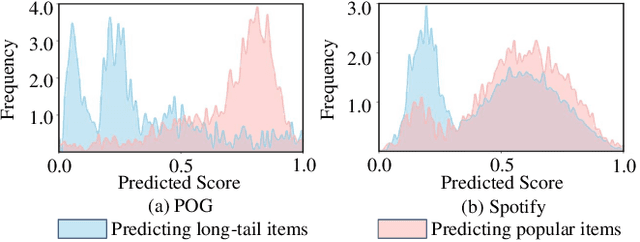
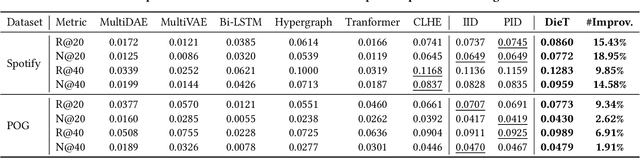
Abstract:Product bundling aims to organize a set of thematically related items into a combined bundle for shipment facilitation and item promotion. To increase the exposure of fresh or overstocked products, sellers typically bundle these items with popular products for inventory clearance. This specific task can be formulated as a long-tail product bundling scenario, which leverages the user-item interactions to define the popularity of each item. The inherent popularity bias in the pre-extracted user feedback features and the insufficient utilization of other popularity-independent knowledge may force the conventional bundling methods to find more popular items, thereby struggling with this long-tail bundling scenario. Through intuitive and empirical analysis, we navigate the core solution for this challenge, which is maximally mining the popularity-free features and effectively incorporating them into the bundling process. To achieve this, we propose a Distilled Modality-Oriented Knowledge Transfer framework (DieT) to effectively counter the popularity bias misintroduced by the user feedback features and adhere to the original intent behind the real-world bundling behaviors. Specifically, DieT first proposes the Popularity-free Collaborative Distribution Modeling module (PCD) to capture the popularity-independent information from the bundle-item view, which is proven most effective in the long-tail bundling scenario to enable the directional information transfer. With the tailored Unbiased Bundle-aware Knowledge Transferring module (UBT), DieT can highlight the significance of popularity-free features while mitigating the negative effects of user feedback features in the long-tail scenario via the knowledge distillation paradigm. Extensive experiments on two real-world datasets demonstrate the superiority of DieT over a list of SOTA methods in the long-tail bundling scenario.
MAC: A Benchmark for Multiple Attributes Compositional Zero-Shot Learning
Jun 18, 2024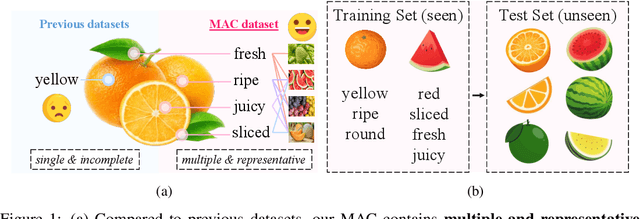

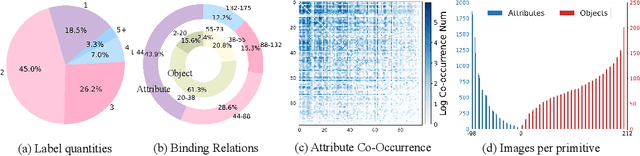
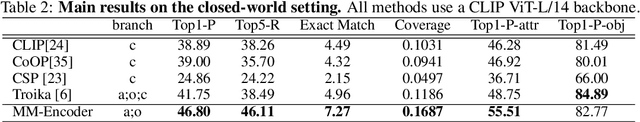
Abstract:Compositional Zero-Shot Learning (CZSL) aims to learn semantic primitives (attributes and objects) from seen compositions and recognize unseen attribute-object compositions. Existing CZSL datasets focus on single attributes, neglecting the fact that objects naturally exhibit multiple interrelated attributes. Real-world objects often possess multiple interrelated attributes, and current datasets' narrow attribute scope and single attribute labeling introduce annotation biases, undermining model performance and evaluation. To address these limitations, we introduce the Multi-Attribute Composition (MAC) dataset, encompassing 18,217 images and 11,067 compositions with comprehensive, representative, and diverse attribute annotations. MAC includes an average of 30.2 attributes per object and 65.4 objects per attribute, facilitating better multi-attribute composition predictions. Our dataset supports deeper semantic understanding and higher-order attribute associations, providing a more realistic and challenging benchmark for the CZSL task. We also develop solutions for multi-attribute compositional learning and propose the MM-encoder to disentangling the attributes and objects.
SimFair: Physics-Guided Fairness-Aware Learning with Simulation Models
Feb 05, 2024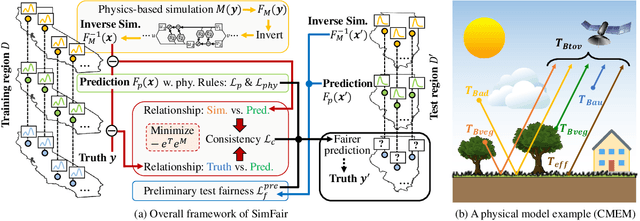
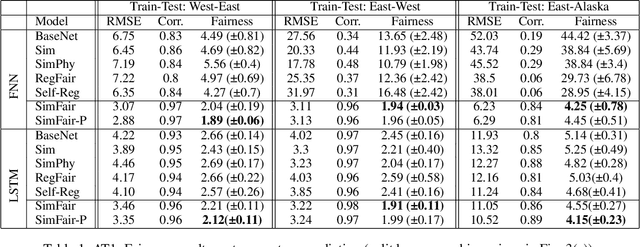
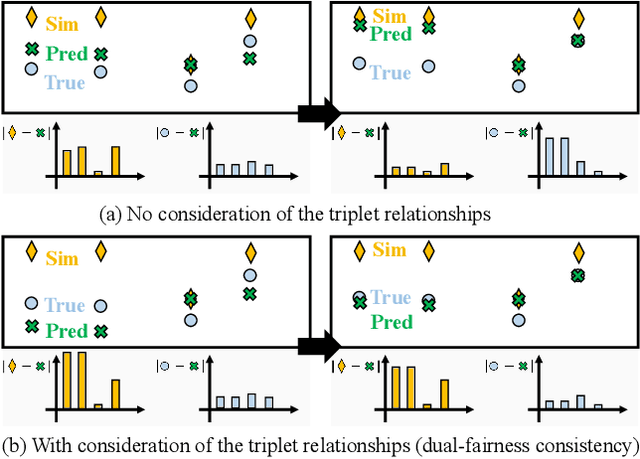
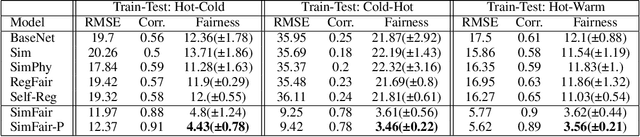
Abstract:Fairness-awareness has emerged as an essential building block for the responsible use of artificial intelligence in real applications. In many cases, inequity in performance is due to the change in distribution over different regions. While techniques have been developed to improve the transferability of fairness, a solution to the problem is not always feasible with no samples from the new regions, which is a bottleneck for pure data-driven attempts. Fortunately, physics-based mechanistic models have been studied for many problems with major social impacts. We propose SimFair, a physics-guided fairness-aware learning framework, which bridges the data limitation by integrating physical-rule-based simulation and inverse modeling into the training design. Using temperature prediction as an example, we demonstrate the effectiveness of the proposed SimFair in fairness preservation.
Deep Radon Prior: A Fully Unsupervised Framework for Sparse-View CT Reconstruction
Dec 30, 2023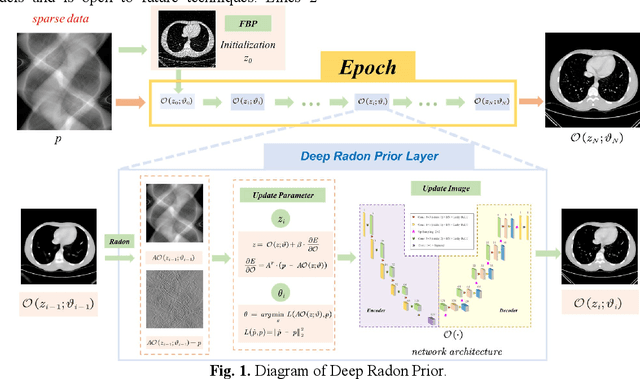
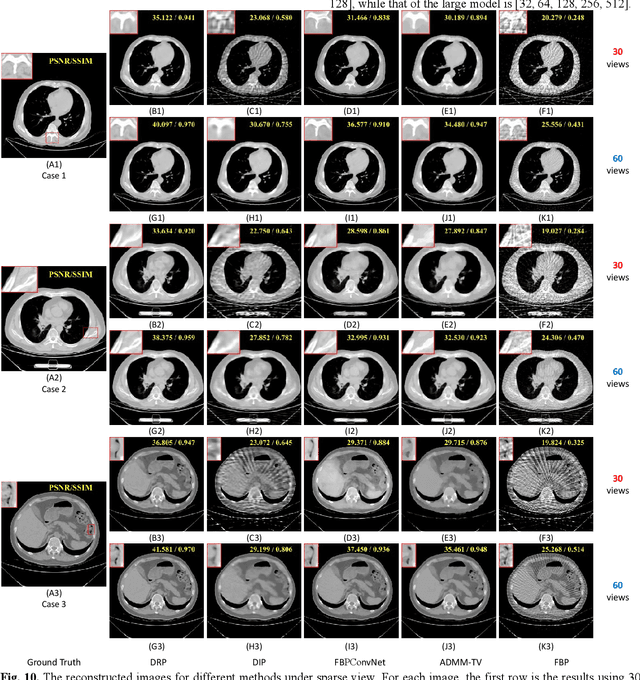
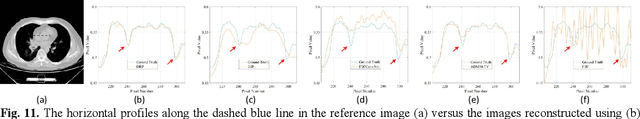
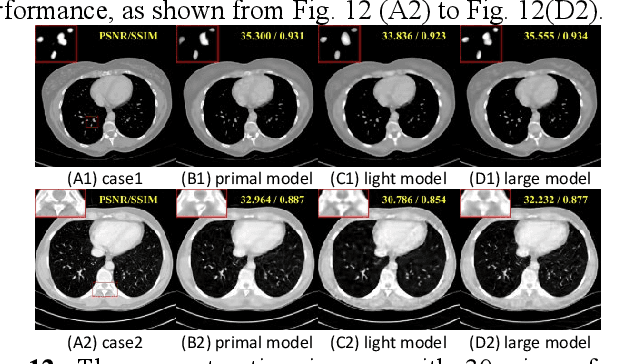
Abstract:Although sparse-view computed tomography (CT) has significantly reduced radiation dose, it also introduces severe artifacts which degrade the image quality. In recent years, deep learning-based methods for inverse problems have made remarkable progress and have become increasingly popular in CT reconstruction. However, most of these methods suffer several limitations: dependence on high-quality training data, weak interpretability, etc. In this study, we propose a fully unsupervised framework called Deep Radon Prior (DRP), inspired by Deep Image Prior (DIP), to address the aforementioned limitations. DRP introduces a neural network as an implicit prior into the iterative method, thereby realizing cross-domain gradient feedback. During the reconstruction process, the neural network is progressively optimized in multiple stages to narrow the solution space in radon domain for the under-constrained imaging protocol, and the convergence of the proposed method has been discussed in this work. Compared with the popular pre-trained method, the proposed framework requires no dataset and exhibits superior interpretability and generalization ability. The experimental results demonstrate that the proposed method can generate detailed images while effectively suppressing image artifacts.Meanwhile, DRP achieves comparable or better performance than the supervised methods.
ICD-LM: Configuring Vision-Language In-Context Demonstrations by Language Modeling
Dec 22, 2023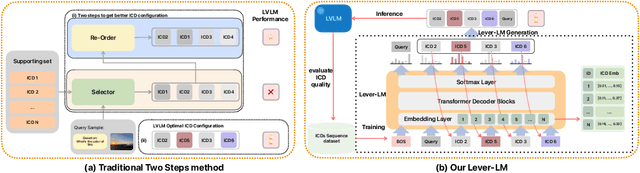
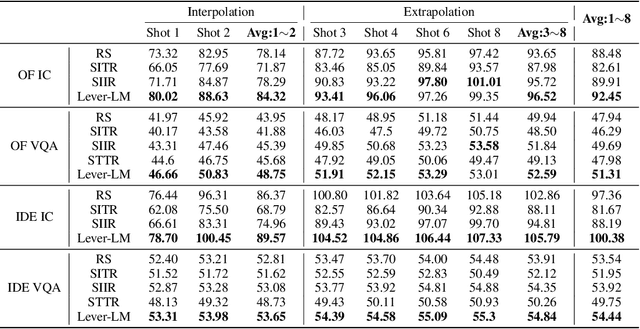
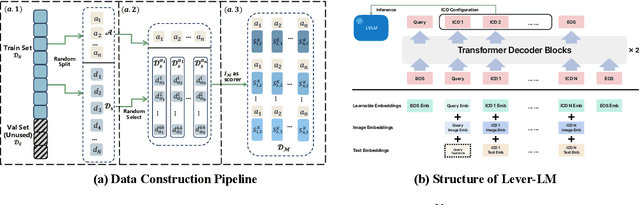
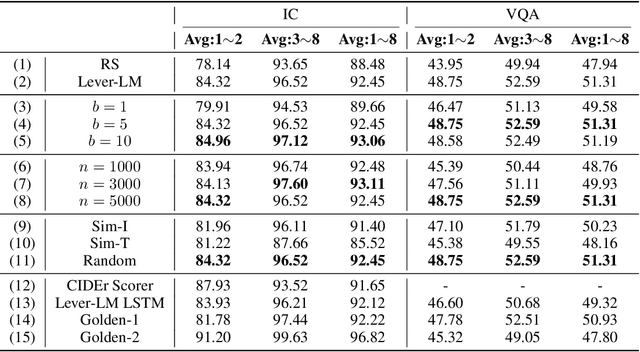
Abstract:This paper studies how to configure powerful In-Context Demonstration (ICD) sequences for a Large Vision-Language Model (LVLM) to solve Vision-Language tasks through In-Context Learning (ICL). After observing that configuring an ICD sequence is a mirror process of composing a sentence, i.e., just as a sentence can be composed word by word via a Language Model, an ICD sequence can also be configured one by one. Consequently, we introduce an ICD Language Model (ICD-LM) specifically designed to generate effective ICD sequences. This involves creating a dataset of hand-crafted ICD sequences for various query samples and using it to train the ICD-LM. Our approach, diverging from traditional methods in NLP that select and order ICDs separately, enables to simultaneously learn how to select and order ICDs, enhancing the effect of the sequences. Moreover, during data construction, we use the LVLM intended for ICL implementation to validate the strength of each ICD sequence, resulting in a model-specific dataset and the ICD-LM trained by this dataset is also model-specific. We validate our methodology through experiments in Visual Question Answering and Image Captioning, confirming the viability of using a Language Model for ICD configuration. Our comprehensive ablation studies further explore the impact of various dataset construction and ICD-LM development settings on the outcomes. The code is given in https://github.com/ForJadeForest/ICD-LM.
Multi-label classification for biomedical literature: an overview of the BioCreative VII LitCovid Track for COVID-19 literature topic annotations
Apr 20, 2022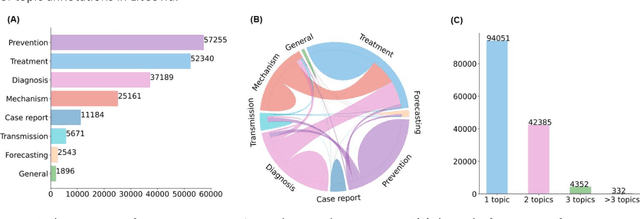
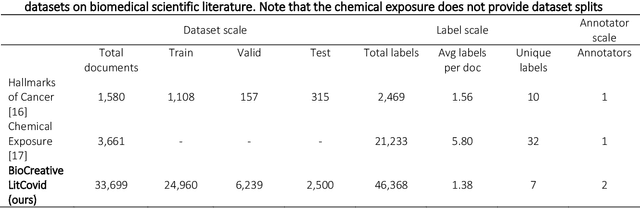
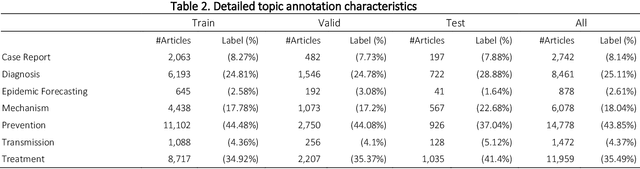
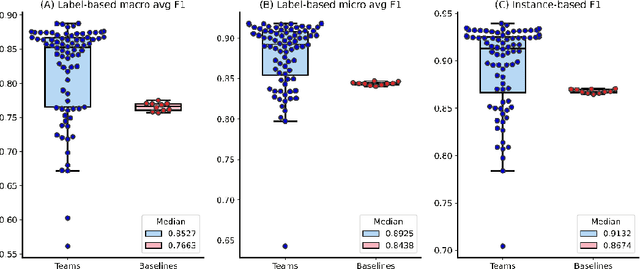
Abstract:The COVID-19 pandemic has been severely impacting global society since December 2019. Massive research has been undertaken to understand the characteristics of the virus and design vaccines and drugs. The related findings have been reported in biomedical literature at a rate of about 10,000 articles on COVID-19 per month. Such rapid growth significantly challenges manual curation and interpretation. For instance, LitCovid is a literature database of COVID-19-related articles in PubMed, which has accumulated more than 200,000 articles with millions of accesses each month by users worldwide. One primary curation task is to assign up to eight topics (e.g., Diagnosis and Treatment) to the articles in LitCovid. Despite the continuing advances in biomedical text mining methods, few have been dedicated to topic annotations in COVID-19 literature. To close the gap, we organized the BioCreative LitCovid track to call for a community effort to tackle automated topic annotation for COVID-19 literature. The BioCreative LitCovid dataset, consisting of over 30,000 articles with manually reviewed topics, was created for training and testing. It is one of the largest multilabel classification datasets in biomedical scientific literature. 19 teams worldwide participated and made 80 submissions in total. Most teams used hybrid systems based on transformers. The highest performing submissions achieved 0.8875, 0.9181, and 0.9394 for macro F1-score, micro F1-score, and instance-based F1-score, respectively. The level of participation and results demonstrate a successful track and help close the gap between dataset curation and method development. The dataset is publicly available via https://ftp.ncbi.nlm.nih.gov/pub/lu/LitCovid/biocreative/ for benchmarking and further development.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge