Zhe He
HARDMath2: A Benchmark for Applied Mathematics Built by Students as Part of a Graduate Class
May 17, 2025Abstract:Large language models (LLMs) have shown remarkable progress in mathematical problem-solving, but evaluation has largely focused on problems that have exact analytical solutions or involve formal proofs, often overlooking approximation-based problems ubiquitous in applied science and engineering. To fill this gap, we build on prior work and present HARDMath2, a dataset of 211 original problems covering the core topics in an introductory graduate applied math class, including boundary-layer analysis, WKB methods, asymptotic solutions of nonlinear partial differential equations, and the asymptotics of oscillatory integrals. This dataset was designed and verified by the students and instructors of a core graduate applied mathematics course at Harvard. We build the dataset through a novel collaborative environment that challenges students to write and refine difficult problems consistent with the class syllabus, peer-validate solutions, test different models, and automatically check LLM-generated solutions against their own answers and numerical ground truths. Evaluation results show that leading frontier models still struggle with many of the problems in the dataset, highlighting a gap in the mathematical reasoning skills of current LLMs. Importantly, students identified strategies to create increasingly difficult problems by interacting with the models and exploiting common failure modes. This back-and-forth with the models not only resulted in a richer and more challenging benchmark but also led to qualitative improvements in the students' understanding of the course material, which is increasingly important as we enter an age where state-of-the-art language models can solve many challenging problems across a wide domain of fields.
POPGym Arcade: Parallel Pixelated POMDPs
Mar 04, 2025Abstract:We introduce POPGym Arcade, a benchmark consisting of 7 pixel-based environments each with three difficulties, utilizing a single observation and action space. Each environment offers both fully observable and partially observable variants, enabling counterfactual studies on partial observability. POPGym Arcade utilizes JIT compilation on hardware accelerators to achieve substantial speedups over CPU-bound environments. Moreover, this enables Podracer-style architectures to further increase hardware utilization and training speed. We evaluate memory models on our environments using a Podracer variant of Q learning, and examine the results. Finally, we generate memory saliency maps, uncovering how memories propagate through policies. Our library is available at https://github.com/bolt-research/popgym_arcade.
Evaluating the Impact of Lab Test Results on Large Language Models Generated Differential Diagnoses from Clinical Case Vignettes
Nov 01, 2024



Abstract:Differential diagnosis is crucial for medicine as it helps healthcare providers systematically distinguish between conditions that share similar symptoms. This study assesses the impact of lab test results on differential diagnoses (DDx) made by large language models (LLMs). Clinical vignettes from 50 case reports from PubMed Central were created incorporating patient demographics, symptoms, and lab results. Five LLMs GPT-4, GPT-3.5, Llama-2-70b, Claude-2, and Mixtral-8x7B were tested to generate Top 10, Top 5, and Top 1 DDx with and without lab data. A comprehensive evaluation involving GPT-4, a knowledge graph, and clinicians was conducted. GPT-4 performed best, achieving 55% accuracy for Top 1 diagnoses and 60% for Top 10 with lab data, with lenient accuracy up to 80%. Lab results significantly improved accuracy, with GPT-4 and Mixtral excelling, though exact match rates were low. Lab tests, including liver function, metabolic/toxicology panels, and serology/immune tests, were generally interpreted correctly by LLMs for differential diagnosis.
Demystifying Large Language Models for Medicine: A Primer
Oct 24, 2024



Abstract:Large language models (LLMs) represent a transformative class of AI tools capable of revolutionizing various aspects of healthcare by generating human-like responses across diverse contexts and adapting to novel tasks following human instructions. Their potential application spans a broad range of medical tasks, such as clinical documentation, matching patients to clinical trials, and answering medical questions. In this primer paper, we propose an actionable guideline to help healthcare professionals more efficiently utilize LLMs in their work, along with a set of best practices. This approach consists of several main phases, including formulating the task, choosing LLMs, prompt engineering, fine-tuning, and deployment. We start with the discussion of critical considerations in identifying healthcare tasks that align with the core capabilities of LLMs and selecting models based on the selected task and data, performance requirements, and model interface. We then review the strategies, such as prompt engineering and fine-tuning, to adapt standard LLMs to specialized medical tasks. Deployment considerations, including regulatory compliance, ethical guidelines, and continuous monitoring for fairness and bias, are also discussed. By providing a structured step-by-step methodology, this tutorial aims to equip healthcare professionals with the tools necessary to effectively integrate LLMs into clinical practice, ensuring that these powerful technologies are applied in a safe, reliable, and impactful manner.
Time Matters: Examine Temporal Effects on Biomedical Language Models
Jul 24, 2024



Abstract:Time roots in applying language models for biomedical applications: models are trained on historical data and will be deployed for new or future data, which may vary from training data. While increasing biomedical tasks have employed state-of-the-art language models, there are very few studies have examined temporal effects on biomedical models when data usually shifts across development and deployment. This study fills the gap by statistically probing relations between language model performance and data shifts across three biomedical tasks. We deploy diverse metrics to evaluate model performance, distance methods to measure data drifts, and statistical methods to quantify temporal effects on biomedical language models. Our study shows that time matters for deploying biomedical language models, while the degree of performance degradation varies by biomedical tasks and statistical quantification approaches. We believe this study can establish a solid benchmark to evaluate and assess temporal effects on deploying biomedical language models.
AgentMD: Empowering Language Agents for Risk Prediction with Large-Scale Clinical Tool Learning
Feb 20, 2024Abstract:Clinical calculators play a vital role in healthcare by offering accurate evidence-based predictions for various purposes such as prognosis. Nevertheless, their widespread utilization is frequently hindered by usability challenges, poor dissemination, and restricted functionality. Augmenting large language models with extensive collections of clinical calculators presents an opportunity to overcome these obstacles and improve workflow efficiency, but the scalability of the manual curation process poses a significant challenge. In response, we introduce AgentMD, a novel language agent capable of curating and applying clinical calculators across various clinical contexts. Using the published literature, AgentMD has automatically curated a collection of 2,164 diverse clinical calculators with executable functions and structured documentation, collectively named RiskCalcs. Manual evaluations show that RiskCalcs tools achieve an accuracy of over 80% on three quality metrics. At inference time, AgentMD can automatically select and apply the relevant RiskCalcs tools given any patient description. On the newly established RiskQA benchmark, AgentMD significantly outperforms chain-of-thought prompting with GPT-4 (87.7% vs. 40.9% in accuracy). Additionally, we also applied AgentMD to real-world clinical notes for analyzing both population-level and risk-level patient characteristics. In summary, our study illustrates the utility of language agents augmented with clinical calculators for healthcare analytics and patient care.
Quality of Answers of Generative Large Language Models vs Peer Patients for Interpreting Lab Test Results for Lay Patients: Evaluation Study
Jan 23, 2024Abstract:Lab results are often confusing and hard to understand. Large language models (LLMs) such as ChatGPT have opened a promising avenue for patients to get their questions answered. We aim to assess the feasibility of using LLMs to generate relevant, accurate, helpful, and unharmful responses to lab test-related questions asked by patients and to identify potential issues that can be mitigated with augmentation approaches. We first collected lab test results related question and answer data from Yahoo! Answers and selected 53 QA pairs for this study. Using the LangChain framework and ChatGPT web portal, we generated responses to the 53 questions from four LLMs including GPT-4, Meta LLaMA 2, MedAlpaca, and ORCA_mini. We first assessed the similarity of their answers using standard QA similarity-based evaluation metrics including ROUGE, BLEU, METEOR, BERTScore. We also utilized an LLM-based evaluator to judge whether a target model has higher quality in terms of relevance, correctness, helpfulness, and safety than the baseline model. Finally, we performed a manual evaluation with medical experts for all the responses to seven selected questions on the same four aspects. The results of Win Rate and medical expert evaluation both showed that GPT-4's responses achieved better scores than all the other LLM responses and human responses on all four aspects (relevance, correctness, helpfulness, and safety). However, LLM responses occasionally also suffer from a lack of interpretation in one's medical context, incorrect statements, and lack of references. We find that compared to other three LLMs and human answer from the Q&A website, GPT-4's responses are more accurate, helpful, relevant, and safer. However, there are cases which GPT-4 responses are inaccurate and not individualized. We identified a number of ways to improve the quality of LLM responses.
Zero-shot Learning with Minimum Instruction to Extract Social Determinants and Family History from Clinical Notes using GPT Model
Sep 13, 2023Abstract:Demographics, Social determinants of health, and family history documented in the unstructured text within the electronic health records are increasingly being studied to understand how this information can be utilized with the structured data to improve healthcare outcomes. After the GPT models were released, many studies have applied GPT models to extract this information from the narrative clinical notes. Different from the existing work, our research focuses on investigating the zero-shot learning on extracting this information together by providing minimum information to the GPT model. We utilize de-identified real-world clinical notes annotated for demographics, various social determinants, and family history information. Given that the GPT model might provide text different from the text in the original data, we explore two sets of evaluation metrics, including the traditional NER evaluation metrics and semantic similarity evaluation metrics, to completely understand the performance. Our results show that the GPT-3.5 method achieved an average of 0.975 F1 on demographics extraction, 0.615 F1 on social determinants extraction, and 0.722 F1 on family history extraction. We believe these results can be further improved through model fine-tuning or few-shots learning. Through the case studies, we also identified the limitations of the GPT models, which need to be addressed in future research.
Can Attention Be Used to Explain EHR-Based Mortality Prediction Tasks: A Case Study on Hemorrhagic Stroke
Aug 04, 2023



Abstract:Stroke is a significant cause of mortality and morbidity, necessitating early predictive strategies to minimize risks. Traditional methods for evaluating patients, such as Acute Physiology and Chronic Health Evaluation (APACHE II, IV) and Simplified Acute Physiology Score III (SAPS III), have limited accuracy and interpretability. This paper proposes a novel approach: an interpretable, attention-based transformer model for early stroke mortality prediction. This model seeks to address the limitations of previous predictive models, providing both interpretability (providing clear, understandable explanations of the model) and fidelity (giving a truthful explanation of the model's dynamics from input to output). Furthermore, the study explores and compares fidelity and interpretability scores using Shapley values and attention-based scores to improve model explainability. The research objectives include designing an interpretable attention-based transformer model, evaluating its performance compared to existing models, and providing feature importance derived from the model.
Improving Primary Healthcare Workflow Using Extreme Summarization of Scientific Literature Based on Generative AI
Jul 24, 2023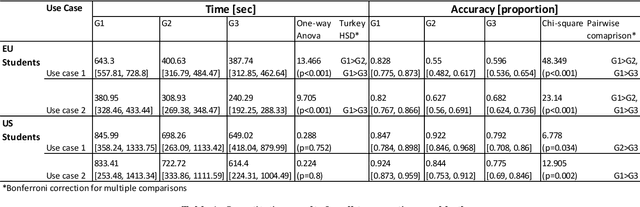
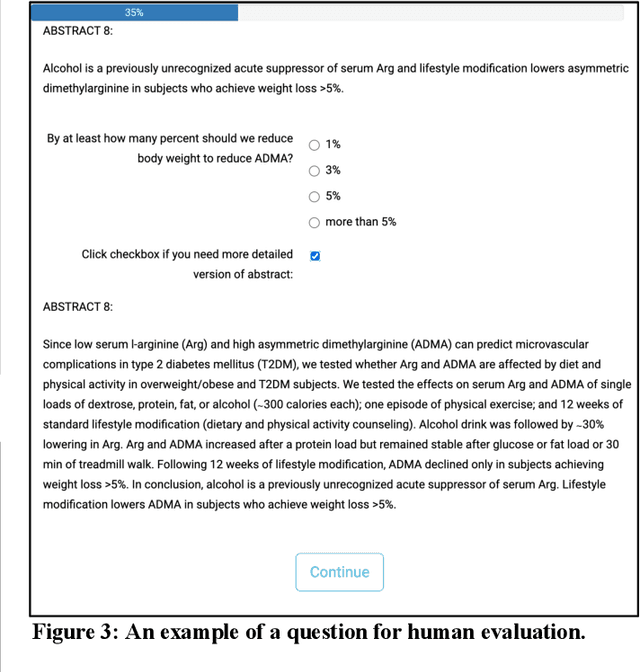
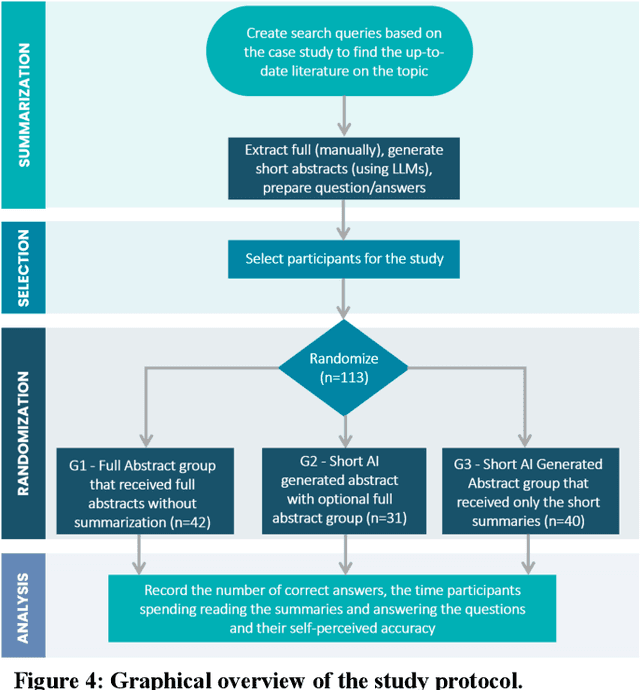
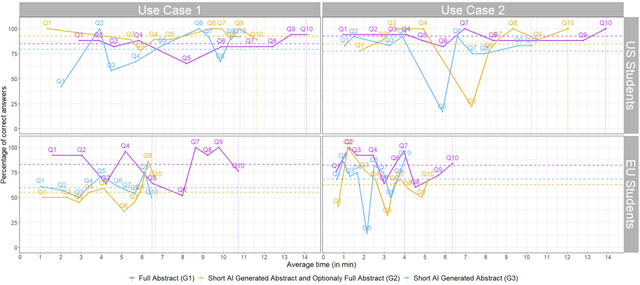
Abstract:Primary care professionals struggle to keep up to date with the latest scientific literature critical in guiding evidence-based practice related to their daily work. To help solve the above-mentioned problem, we employed generative artificial intelligence techniques based on large-scale language models to summarize abstracts of scientific papers. Our objective is to investigate the potential of generative artificial intelligence in diminishing the cognitive load experienced by practitioners, thus exploring its ability to alleviate mental effort and burden. The study participants were provided with two use cases related to preventive care and behavior change, simulating a search for new scientific literature. The study included 113 university students from Slovenia and the United States randomized into three distinct study groups. The first group was assigned to the full abstracts. The second group was assigned to the short abstracts generated by AI. The third group had the option to select a full abstract in addition to the AI-generated short summary. Each use case study included ten retrieved abstracts. Our research demonstrates that the use of generative AI for literature review is efficient and effective. The time needed to answer questions related to the content of abstracts was significantly lower in groups two and three compared to the first group using full abstracts. The results, however, also show significantly lower accuracy in extracted knowledge in cases where full abstract was not available. Such a disruptive technology could significantly reduce the time required for healthcare professionals to keep up with the most recent scientific literature; nevertheless, further developments are needed to help them comprehend the knowledge accurately.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge