Prithwish Chakraborty
MetaExplainer: A Framework to Generate Multi-Type User-Centered Explanations for AI Systems
Aug 01, 2025Abstract:Explanations are crucial for building trustworthy AI systems, but a gap often exists between the explanations provided by models and those needed by users. To address this gap, we introduce MetaExplainer, a neuro-symbolic framework designed to generate user-centered explanations. Our approach employs a three-stage process: first, we decompose user questions into machine-readable formats using state-of-the-art large language models (LLM); second, we delegate the task of generating system recommendations to model explainer methods; and finally, we synthesize natural language explanations that summarize the explainer outputs. Throughout this process, we utilize an Explanation Ontology to guide the language models and explainer methods. By leveraging LLMs and a structured approach to explanation generation, MetaExplainer aims to enhance the interpretability and trustworthiness of AI systems across various applications, providing users with tailored, question-driven explanations that better meet their needs. Comprehensive evaluations of MetaExplainer demonstrate a step towards evaluating and utilizing current state-of-the-art explanation frameworks. Our results show high performance across all stages, with a 59.06% F1-score in question reframing, 70% faithfulness in model explanations, and 67% context-utilization in natural language synthesis. User studies corroborate these findings, highlighting the creativity and comprehensiveness of generated explanations. Tested on the Diabetes (PIMA Indian) tabular dataset, MetaExplainer supports diverse explanation types, including Contrastive, Counterfactual, Rationale, Case-Based, and Data explanations. The framework's versatility and traceability from using ontology to guide LLMs suggest broad applicability beyond the tested scenarios, positioning MetaExplainer as a promising tool for enhancing AI explainability across various domains.
Improving Primary Healthcare Workflow Using Extreme Summarization of Scientific Literature Based on Generative AI
Jul 24, 2023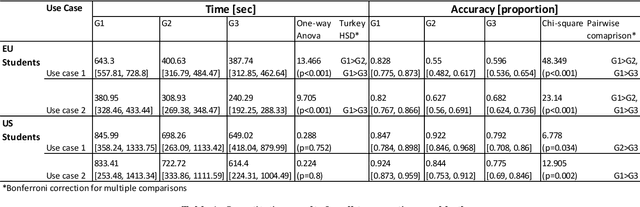
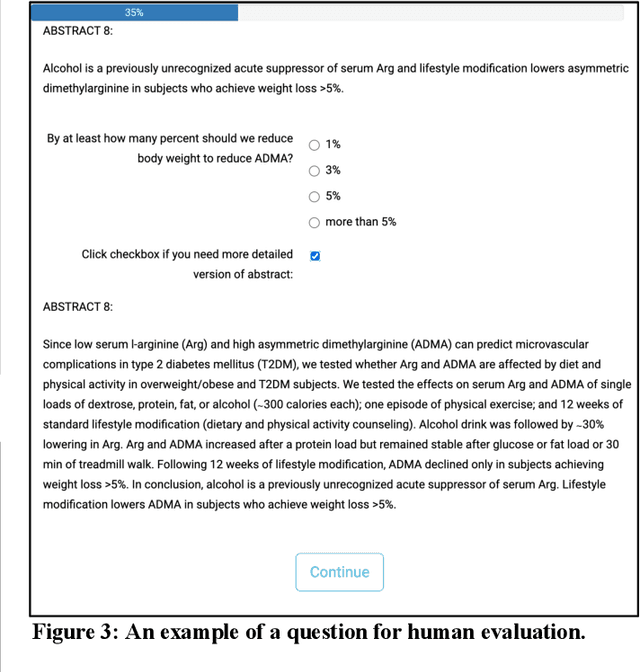
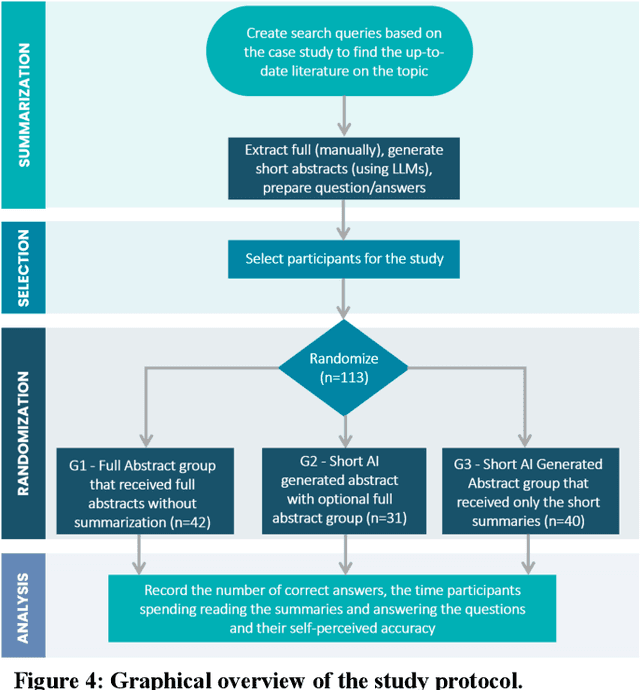
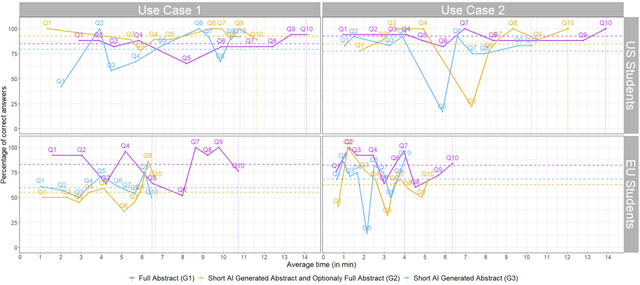
Abstract:Primary care professionals struggle to keep up to date with the latest scientific literature critical in guiding evidence-based practice related to their daily work. To help solve the above-mentioned problem, we employed generative artificial intelligence techniques based on large-scale language models to summarize abstracts of scientific papers. Our objective is to investigate the potential of generative artificial intelligence in diminishing the cognitive load experienced by practitioners, thus exploring its ability to alleviate mental effort and burden. The study participants were provided with two use cases related to preventive care and behavior change, simulating a search for new scientific literature. The study included 113 university students from Slovenia and the United States randomized into three distinct study groups. The first group was assigned to the full abstracts. The second group was assigned to the short abstracts generated by AI. The third group had the option to select a full abstract in addition to the AI-generated short summary. Each use case study included ten retrieved abstracts. Our research demonstrates that the use of generative AI for literature review is efficient and effective. The time needed to answer questions related to the content of abstracts was significantly lower in groups two and three compared to the first group using full abstracts. The results, however, also show significantly lower accuracy in extracted knowledge in cases where full abstract was not available. Such a disruptive technology could significantly reduce the time required for healthcare professionals to keep up with the most recent scientific literature; nevertheless, further developments are needed to help them comprehend the knowledge accurately.
Informing clinical assessment by contextualizing post-hoc explanations of risk prediction models in type-2 diabetes
Feb 11, 2023Abstract:Medical experts may use Artificial Intelligence (AI) systems with greater trust if these are supported by contextual explanations that let the practitioner connect system inferences to their context of use. However, their importance in improving model usage and understanding has not been extensively studied. Hence, we consider a comorbidity risk prediction scenario and focus on contexts regarding the patients clinical state, AI predictions about their risk of complications, and algorithmic explanations supporting the predictions. We explore how relevant information for such dimensions can be extracted from Medical guidelines to answer typical questions from clinical practitioners. We identify this as a question answering (QA) task and employ several state-of-the-art LLMs to present contexts around risk prediction model inferences and evaluate their acceptability. Finally, we study the benefits of contextual explanations by building an end-to-end AI pipeline including data cohorting, AI risk modeling, post-hoc model explanations, and prototyped a visual dashboard to present the combined insights from different context dimensions and data sources, while predicting and identifying the drivers of risk of Chronic Kidney Disease - a common type-2 diabetes comorbidity. All of these steps were performed in engagement with medical experts, including a final evaluation of the dashboard results by an expert medical panel. We show that LLMs, in particular BERT and SciBERT, can be readily deployed to extract some relevant explanations to support clinical usage. To understand the value-add of the contextual explanations, the expert panel evaluated these regarding actionable insights in the relevant clinical setting. Overall, our paper is one of the first end-to-end analyses identifying the feasibility and benefits of contextual explanations in a real-world clinical use case.
Distillation to Enhance the Portability of Risk Models Across Institutions with Large Patient Claims Database
Jul 06, 2022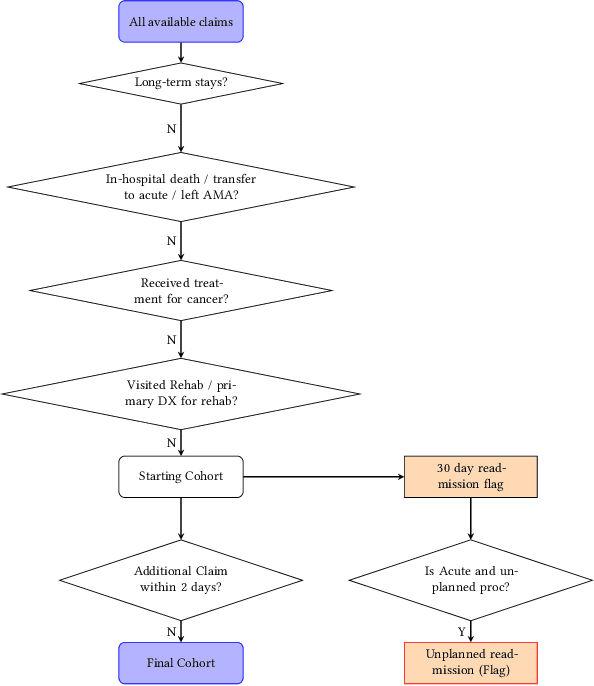
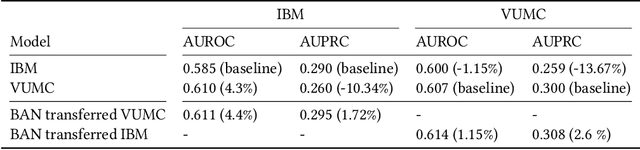
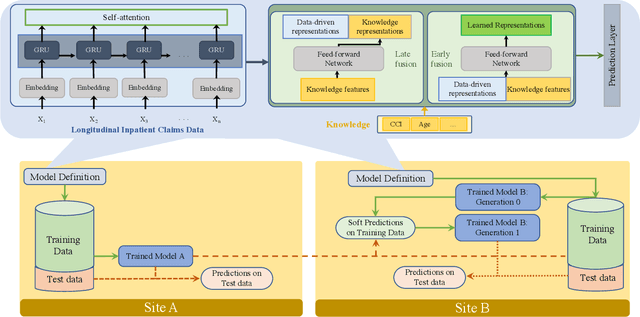

Abstract:Artificial intelligence, and particularly machine learning (ML), is increasingly developed and deployed to support healthcare in a variety of settings. However, clinical decision support (CDS) technologies based on ML need to be portable if they are to be adopted on a broad scale. In this respect, models developed at one institution should be reusable at another. Yet there are numerous examples of portability failure, particularly due to naive application of ML models. Portability failure can lead to suboptimal care and medical errors, which ultimately could prevent the adoption of ML-based CDS in practice. One specific healthcare challenge that could benefit from enhanced portability is the prediction of 30-day readmission risk. Research to date has shown that deep learning models can be effective at modeling such risk. In this work, we investigate the practicality of model portability through a cross-site evaluation of readmission prediction models. To do so, we apply a recurrent neural network, augmented with self-attention and blended with expert features, to build readmission prediction models for two independent large scale claims datasets. We further present a novel transfer learning technique that adapts the well-known method of born-again network (BAN) training. Our experiments show that direct application of ML models trained at one institution and tested at another institution perform worse than models trained and tested at the same institution. We further show that the transfer learning approach based on the BAN produces models that are better than those trained on just a single institution's data. Notably, this improvement is consistent across both sites and occurs after a single retraining, which illustrates the potential for a cheap and general model transfer mechanism of readmission risk prediction.
Leveraging Clinical Context for User-Centered Explainability: A Diabetes Use Case
Jul 15, 2021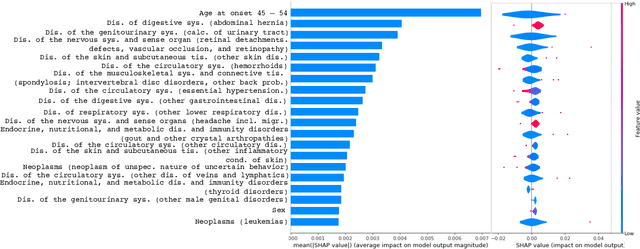
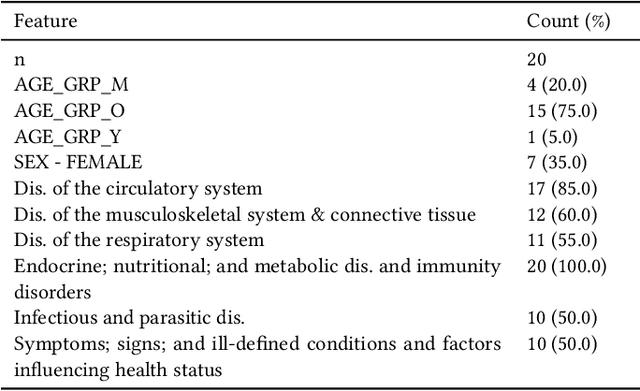
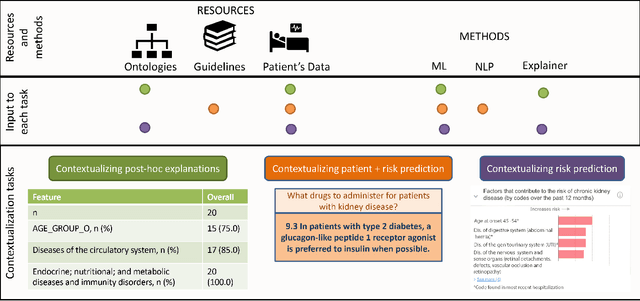
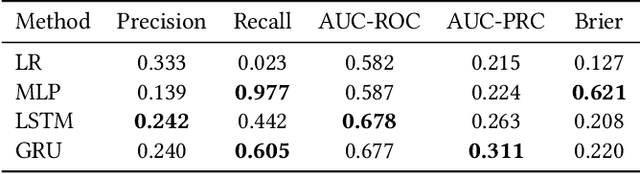
Abstract:Academic advances of AI models in high-precision domains, like healthcare, need to be made explainable in order to enhance real-world adoption. Our past studies and ongoing interactions indicate that medical experts can use AI systems with greater trust if there are ways to connect the model inferences about patients to explanations that are tied back to the context of use. Specifically, risk prediction is a complex problem of diagnostic and interventional importance to clinicians wherein they consult different sources to make decisions. To enable the adoption of the ever improving AI risk prediction models in practice, we have begun to explore techniques to contextualize such models along three dimensions of interest: the patients' clinical state, AI predictions about their risk of complications, and algorithmic explanations supporting the predictions. We validate the importance of these dimensions by implementing a proof-of-concept (POC) in type-2 diabetes (T2DM) use case where we assess the risk of chronic kidney disease (CKD) - a common T2DM comorbidity. Within the POC, we include risk prediction models for CKD, post-hoc explainers of the predictions, and other natural-language modules which operationalize domain knowledge and CPGs to provide context. With primary care physicians (PCP) as our end-users, we present our initial results and clinician feedback in this paper. Our POC approach covers multiple knowledge sources and clinical scenarios, blends knowledge to explain data and predictions to PCPs, and received an enthusiastic response from our medical expert.
Disease Progression Modeling Workbench 360
Jun 24, 2021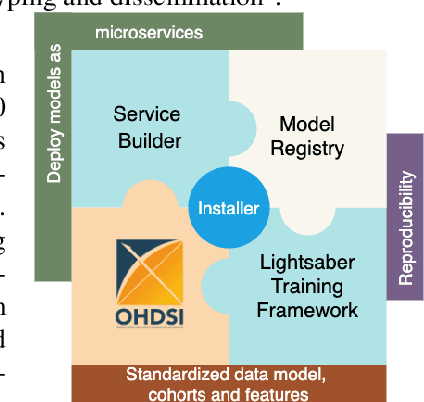
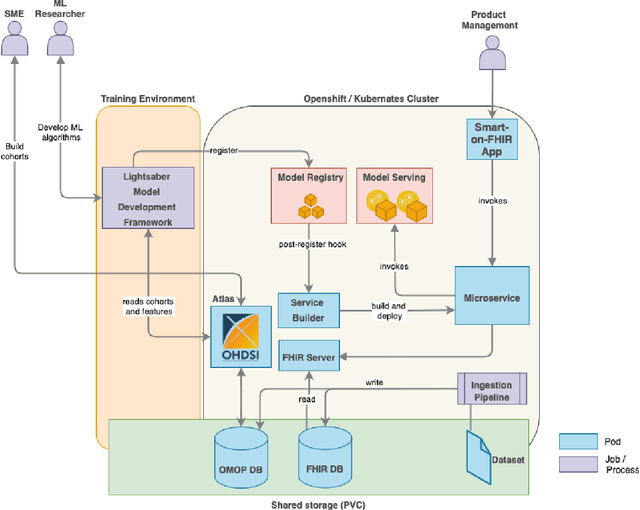
Abstract:In this work we introduce Disease Progression Modeling workbench 360 (DPM360) opensource clinical informatics framework for collaborative research and delivery of healthcare AI. DPM360, when fully developed, will manage the entire modeling life cycle, from data analysis (e.g., cohort identification) to machine learning algorithm development and prototyping. DPM360 augments the advantages of data model standardization and tooling (OMOP-CDM, Athena, ATLAS) provided by the widely-adopted OHDSI initiative with a powerful machine learning training framework, and a mechanism for rapid prototyping through automatic deployment of models as containerized services to a cloud environment.
Collaborative Graph Learning with Auxiliary Text for Temporal Event Prediction in Healthcare
May 16, 2021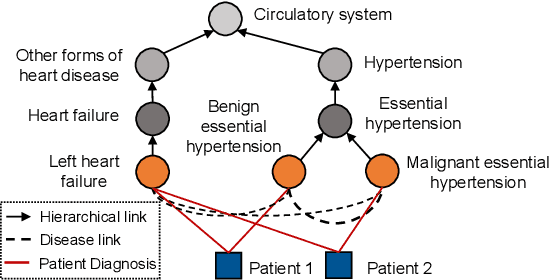

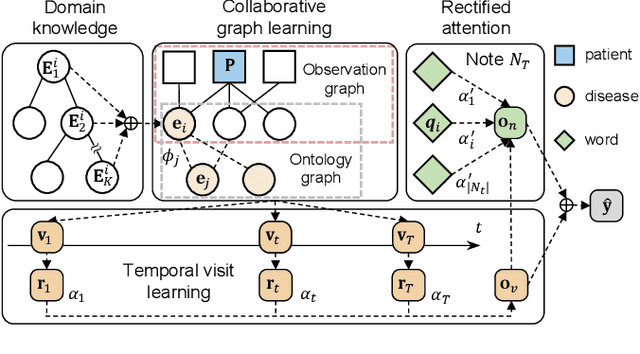
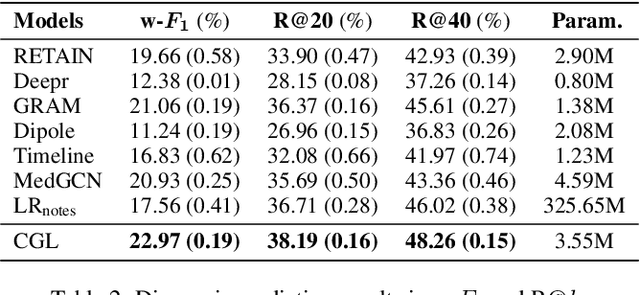
Abstract:Accurate and explainable health event predictions are becoming crucial for healthcare providers to develop care plans for patients. The availability of electronic health records (EHR) has enabled machine learning advances in providing these predictions. However, many deep learning based methods are not satisfactory in solving several key challenges: 1) effectively utilizing disease domain knowledge; 2) collaboratively learning representations of patients and diseases; and 3) incorporating unstructured text. To address these issues, we propose a collaborative graph learning model to explore patient-disease interactions and medical domain knowledge. Our solution is able to capture structural features of both patients and diseases. The proposed model also utilizes unstructured text data by employing an attention regulation strategy and then integrates attentive text features into a sequential learning process. We conduct extensive experiments on two important healthcare problems to show the competitive prediction performance of the proposed method compared with various state-of-the-art models. We also confirm the effectiveness of learned representations and model interpretability by a set of ablation and case studies.
Blending Knowledge in Deep Recurrent Networks for Adverse Event Prediction at Hospital Discharge
Apr 09, 2021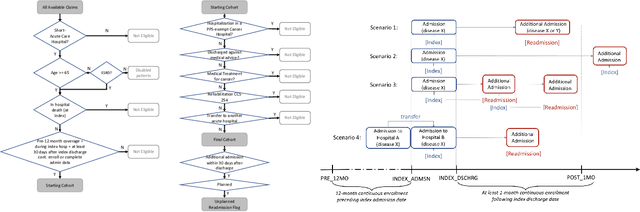
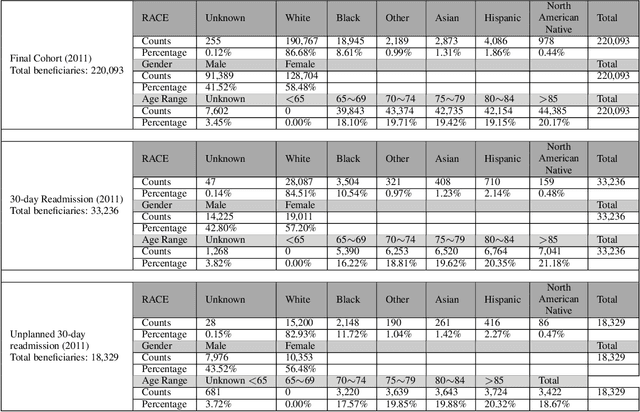
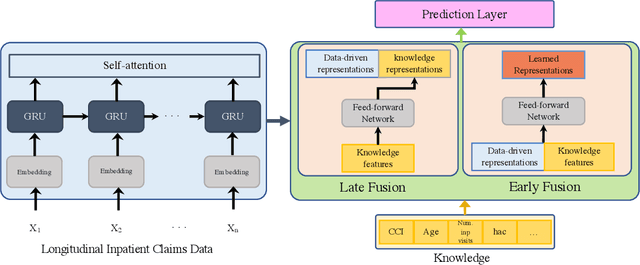

Abstract:Deep learning architectures have an extremely high-capacity for modeling complex data in a wide variety of domains. However, these architectures have been limited in their ability to support complex prediction problems using insurance claims data, such as readmission at 30 days, mainly due to data sparsity issue. Consequently, classical machine learning methods, especially those that embed domain knowledge in handcrafted features, are often on par with, and sometimes outperform, deep learning approaches. In this paper, we illustrate how the potential of deep learning can be achieved by blending domain knowledge within deep learning architectures to predict adverse events at hospital discharge, including readmissions. More specifically, we introduce a learning architecture that fuses a representation of patient data computed by a self-attention based recurrent neural network, with clinically relevant features. We conduct extensive experiments on a large claims dataset and show that the blended method outperforms the standard machine learning approaches.
Phenotypical Ontology Driven Framework for Multi-Task Learning
Sep 04, 2020
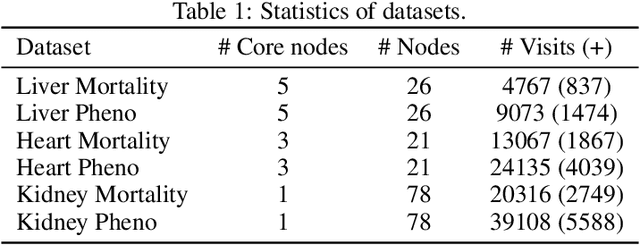
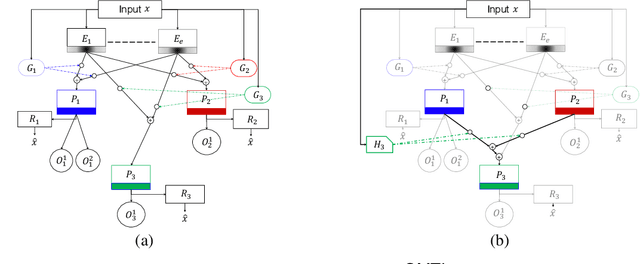
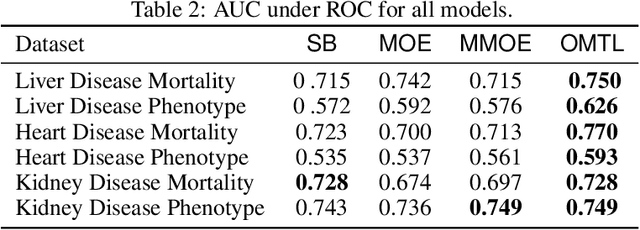
Abstract:Despite the large number of patients in Electronic Health Records (EHRs), the subset of usable data for modeling outcomes of specific phenotypes are often imbalanced and of modest size. This can be attributed to the uneven coverage of medical concepts in EHRs. In this paper, we propose OMTL, an Ontology-driven Multi-Task Learning framework, that is designed to overcome such data limitations. The key contribution of our work is the effective use of knowledge from a predefined well-established medical relationship graph (ontology) to construct a novel deep learning network architecture that mirrors this ontology. It can effectively leverage knowledge from a well-established medical relationship graph (ontology) by constructing a deep learning network architecture that mirrors this graph. This enables common representations to be shared across related phenotypes, and was found to improve the learning performance. The proposed OMTL naturally allows for multitask learning of different phenotypes on distinct predictive tasks. These phenotypes are tied together by their semantic distance according to the external medical ontology. Using the publicly available MIMIC-III database, we evaluate OMTL and demonstrate its efficacy on several real patient outcome predictions over state-of-the-art multi-task learning schemes.
A Canonical Architecture For Predictive Analytics on Longitudinal Patient Records
Jul 24, 2020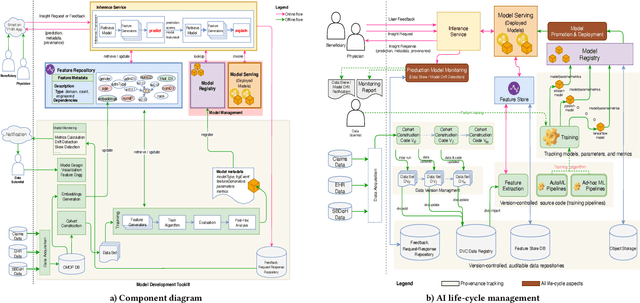
Abstract:Many institutions within the healthcare ecosystem are making significant investments in AI technologies to optimize their business operations at lower cost with improved patient outcomes. Despite the hype with AI, the full realization of this potential is seriously hindered by several systemic problems, including data privacy, security, bias, fairness, and explainability. In this paper, we propose a novel canonical architecture for the development of AI models in healthcare that addresses these challenges. This system enables the creation and management of AI predictive models throughout all the phases of their life cycle, including data ingestion, model building, and model promotion in production environments. This paper describes this architecture in detail, along with a qualitative evaluation of our experience of using it on real world problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge