Mohamed Ghalwash
Informing clinical assessment by contextualizing post-hoc explanations of risk prediction models in type-2 diabetes
Feb 11, 2023Abstract:Medical experts may use Artificial Intelligence (AI) systems with greater trust if these are supported by contextual explanations that let the practitioner connect system inferences to their context of use. However, their importance in improving model usage and understanding has not been extensively studied. Hence, we consider a comorbidity risk prediction scenario and focus on contexts regarding the patients clinical state, AI predictions about their risk of complications, and algorithmic explanations supporting the predictions. We explore how relevant information for such dimensions can be extracted from Medical guidelines to answer typical questions from clinical practitioners. We identify this as a question answering (QA) task and employ several state-of-the-art LLMs to present contexts around risk prediction model inferences and evaluate their acceptability. Finally, we study the benefits of contextual explanations by building an end-to-end AI pipeline including data cohorting, AI risk modeling, post-hoc model explanations, and prototyped a visual dashboard to present the combined insights from different context dimensions and data sources, while predicting and identifying the drivers of risk of Chronic Kidney Disease - a common type-2 diabetes comorbidity. All of these steps were performed in engagement with medical experts, including a final evaluation of the dashboard results by an expert medical panel. We show that LLMs, in particular BERT and SciBERT, can be readily deployed to extract some relevant explanations to support clinical usage. To understand the value-add of the contextual explanations, the expert panel evaluated these regarding actionable insights in the relevant clinical setting. Overall, our paper is one of the first end-to-end analyses identifying the feasibility and benefits of contextual explanations in a real-world clinical use case.
Leveraging Clinical Context for User-Centered Explainability: A Diabetes Use Case
Jul 15, 2021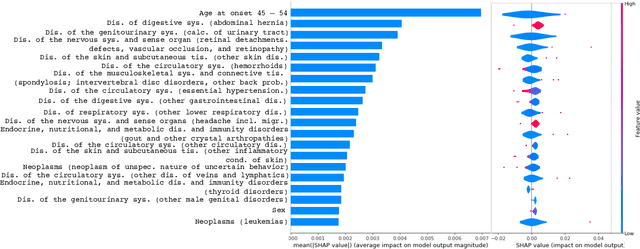
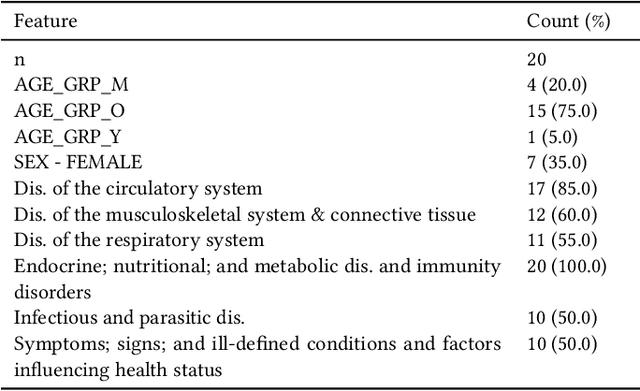
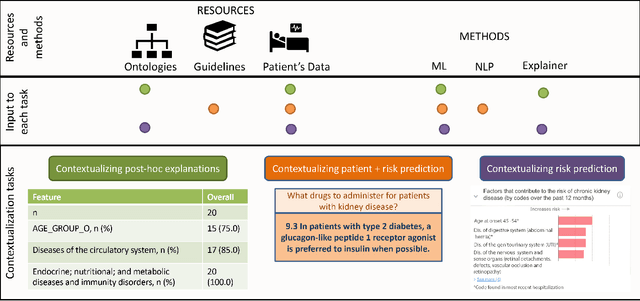
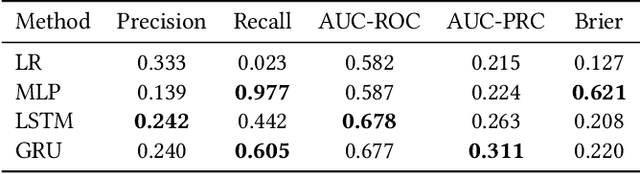
Abstract:Academic advances of AI models in high-precision domains, like healthcare, need to be made explainable in order to enhance real-world adoption. Our past studies and ongoing interactions indicate that medical experts can use AI systems with greater trust if there are ways to connect the model inferences about patients to explanations that are tied back to the context of use. Specifically, risk prediction is a complex problem of diagnostic and interventional importance to clinicians wherein they consult different sources to make decisions. To enable the adoption of the ever improving AI risk prediction models in practice, we have begun to explore techniques to contextualize such models along three dimensions of interest: the patients' clinical state, AI predictions about their risk of complications, and algorithmic explanations supporting the predictions. We validate the importance of these dimensions by implementing a proof-of-concept (POC) in type-2 diabetes (T2DM) use case where we assess the risk of chronic kidney disease (CKD) - a common T2DM comorbidity. Within the POC, we include risk prediction models for CKD, post-hoc explainers of the predictions, and other natural-language modules which operationalize domain knowledge and CPGs to provide context. With primary care physicians (PCP) as our end-users, we present our initial results and clinician feedback in this paper. Our POC approach covers multiple knowledge sources and clinical scenarios, blends knowledge to explain data and predictions to PCPs, and received an enthusiastic response from our medical expert.
Disease Progression Modeling Workbench 360
Jun 24, 2021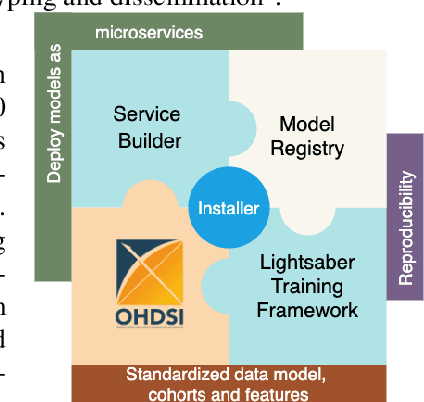
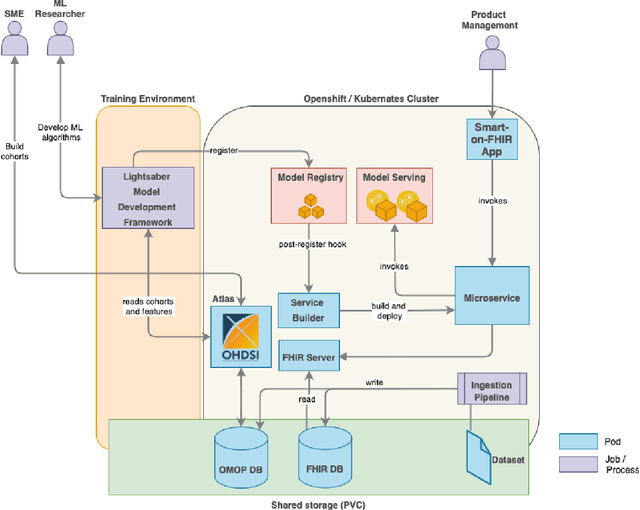
Abstract:In this work we introduce Disease Progression Modeling workbench 360 (DPM360) opensource clinical informatics framework for collaborative research and delivery of healthcare AI. DPM360, when fully developed, will manage the entire modeling life cycle, from data analysis (e.g., cohort identification) to machine learning algorithm development and prototyping. DPM360 augments the advantages of data model standardization and tooling (OMOP-CDM, Athena, ATLAS) provided by the widely-adopted OHDSI initiative with a powerful machine learning training framework, and a mechanism for rapid prototyping through automatic deployment of models as containerized services to a cloud environment.
Blending Knowledge in Deep Recurrent Networks for Adverse Event Prediction at Hospital Discharge
Apr 09, 2021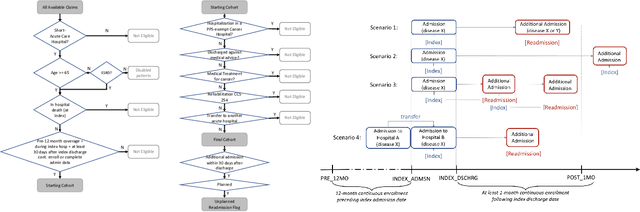
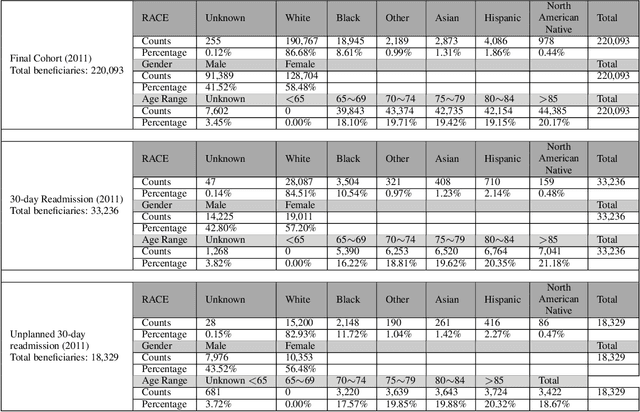
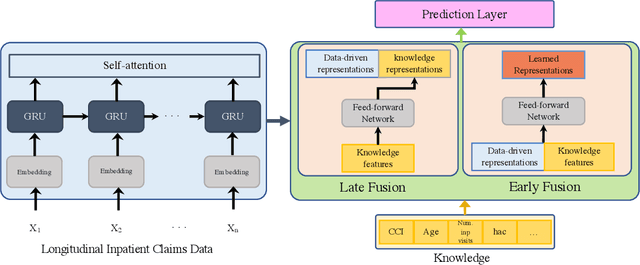

Abstract:Deep learning architectures have an extremely high-capacity for modeling complex data in a wide variety of domains. However, these architectures have been limited in their ability to support complex prediction problems using insurance claims data, such as readmission at 30 days, mainly due to data sparsity issue. Consequently, classical machine learning methods, especially those that embed domain knowledge in handcrafted features, are often on par with, and sometimes outperform, deep learning approaches. In this paper, we illustrate how the potential of deep learning can be achieved by blending domain knowledge within deep learning architectures to predict adverse events at hospital discharge, including readmissions. More specifically, we introduce a learning architecture that fuses a representation of patient data computed by a self-attention based recurrent neural network, with clinically relevant features. We conduct extensive experiments on a large claims dataset and show that the blended method outperforms the standard machine learning approaches.
Phenotypical Ontology Driven Framework for Multi-Task Learning
Sep 04, 2020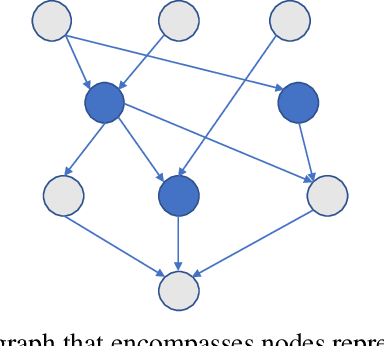
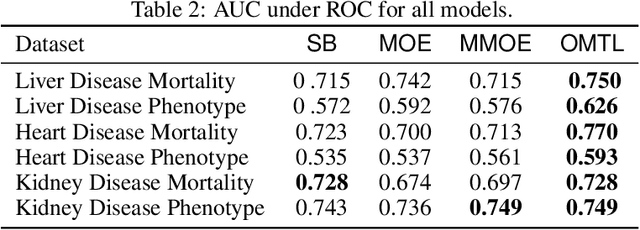
Abstract:Despite the large number of patients in Electronic Health Records (EHRs), the subset of usable data for modeling outcomes of specific phenotypes are often imbalanced and of modest size. This can be attributed to the uneven coverage of medical concepts in EHRs. In this paper, we propose OMTL, an Ontology-driven Multi-Task Learning framework, that is designed to overcome such data limitations. The key contribution of our work is the effective use of knowledge from a predefined well-established medical relationship graph (ontology) to construct a novel deep learning network architecture that mirrors this ontology. It can effectively leverage knowledge from a well-established medical relationship graph (ontology) by constructing a deep learning network architecture that mirrors this graph. This enables common representations to be shared across related phenotypes, and was found to improve the learning performance. The proposed OMTL naturally allows for multitask learning of different phenotypes on distinct predictive tasks. These phenotypes are tied together by their semantic distance according to the external medical ontology. Using the publicly available MIMIC-III database, we evaluate OMTL and demonstrate its efficacy on several real patient outcome predictions over state-of-the-art multi-task learning schemes.
ODVICE: An Ontology-Driven Visual Analytic Tool for Interactive Cohort Extraction
May 13, 2020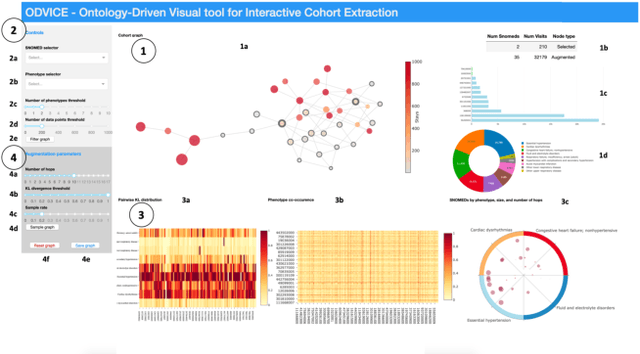

Abstract:Increased availability of electronic health records (EHR) has enabled researchers to study various medical questions. Cohort selection for the hypothesis under investigation is one of the main consideration for EHR analysis. For uncommon diseases, cohorts extracted from EHRs contain very limited number of records - hampering the robustness of any analysis. Data augmentation methods have been successfully applied in other domains to address this issue mainly using simulated records. In this paper, we present ODVICE, a data augmentation framework that leverages the medical concept ontology to systematically augment records using a novel ontologically guided Monte-Carlo graph spanning algorithm. The tool allows end users to specify a small set of interactive controls to control the augmentation process. We analyze the importance of ODVICE by conducting studies on MIMIC-III dataset for two learning tasks. Our results demonstrate the predictive performance of ODVICE augmented cohorts, showing ~30% improvement in area under the curve (AUC) over the non-augmented dataset and other data augmentation strategies.
G-Net: A Deep Learning Approach to G-computation for Counterfactual Outcome Prediction Under Dynamic Treatment Regimes
Mar 23, 2020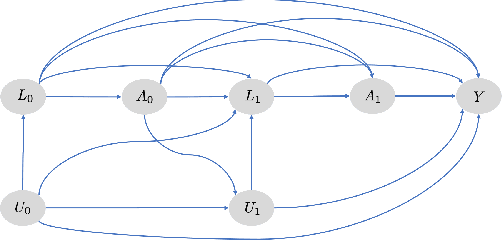
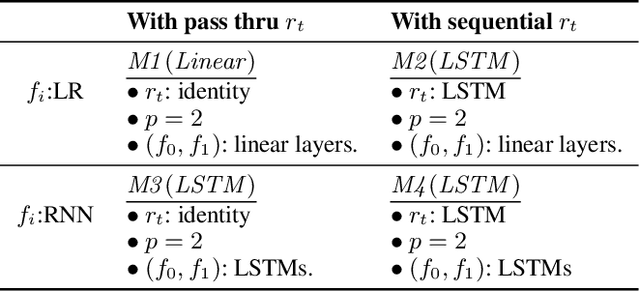
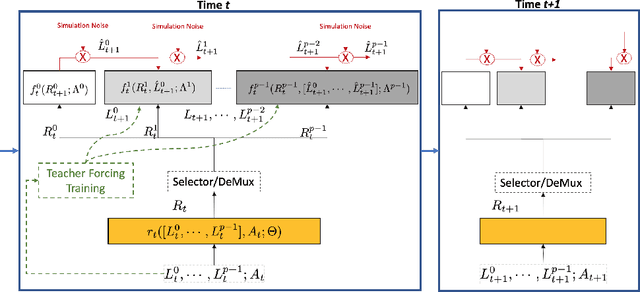
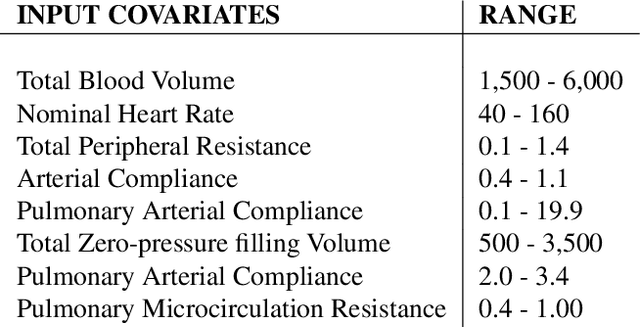
Abstract:Counterfactual prediction is a fundamental task in decision-making. G-computation is a method for estimating expected counterfactual outcomes under dynamic time-varying treatment strategies. Existing G-computation implementations have mostly employed classical regression models with limited capacity to capture complex temporal and nonlinear dependence structures. This paper introduces G-Net, a novel sequential deep learning framework for G-computation that can handle complex time series data while imposing minimal modeling assumptions and provide estimates of individual or population-level time varying treatment effects. We evaluate alternative G-Net implementations using realistically complex temporal simulated data obtained from CVSim, a mechanistic model of the cardiovascular system.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge