Yuqi Si
Hierarchical Transformer Networks for Longitudinal Clinical Document Classification
Apr 17, 2021



Abstract:We present the Hierarchical Transformer Networks for modeling long-term dependencies across clinical notes for the purpose of patient-level prediction. The network is equipped with three levels of Transformer-based encoders to learn progressively from words to sentences, sentences to notes, and finally notes to patients. The first level from word to sentence directly applies a pre-trained BERT model, and the second and third levels both implement a stack of 2-layer encoders before the final patient representation is fed into the classification layer for clinical predictions. Compared to traditional BERT models, our model increases the maximum input length from 512 words to much longer sequences that are appropriate for long sequences of clinical notes. We empirically examine and experiment with different parameters to identify an optimal trade-off given computational resource limits. Our experimental results on the MIMIC-III dataset for different prediction tasks demonstrate that our proposed hierarchical model outperforms previous state-of-the-art hierarchical neural networks.
Generalized and Transferable Patient Language Representation for Phenotyping with Limited Data
Feb 24, 2021



Abstract:The paradigm of representation learning through transfer learning has the potential to greatly enhance clinical natural language processing. In this work, we propose a multi-task pre-training and fine-tuning approach for learning generalized and transferable patient representations from medical language. The model is first pre-trained with different but related high-prevalence phenotypes and further fine-tuned on downstream target tasks. Our main contribution focuses on the impact this technique can have on low-prevalence phenotypes, a challenging task due to the dearth of data. We validate the representation from pre-training, and fine-tune the multi-task pre-trained models on low-prevalence phenotypes including 38 circulatory diseases, 23 respiratory diseases, and 17 genitourinary diseases. We find multi-task pre-training increases learning efficiency and achieves consistently high performance across the majority of phenotypes. Most important, the multi-task pre-training is almost always either the best-performing model or performs tolerably close to the best-performing model, a property we refer to as robust. All these results lead us to conclude that this multi-task transfer learning architecture is a robust approach for developing generalized and transferable patient language representations for numerous phenotypes.
Deep Representation Learning of Patient Data from Electronic Health Records (EHR): A Systematic Review
Oct 06, 2020
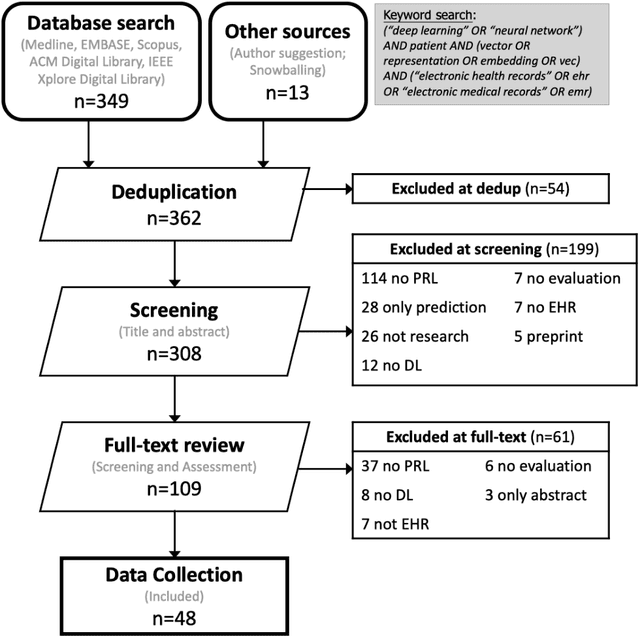
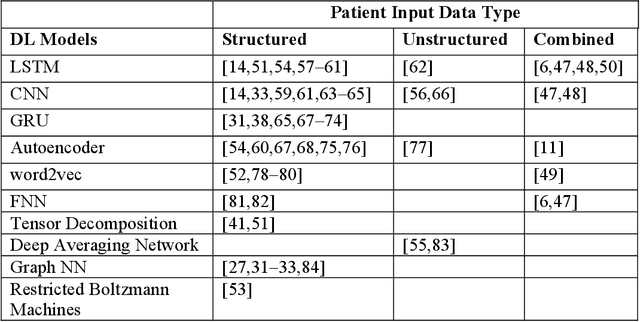
Abstract:Patient representation learning refers to learning a dense mathematical representation of a patient that encodes meaningful information from Electronic Health Records (EHRs). This is generally performed using advanced deep learning methods. This study presents a systematic review of this field and provides both qualitative and quantitative analyses from a methodological perspective. We identified studies developing patient representations from EHRs with deep learning methods from MEDLINE, EMBASE, Scopus, the Association for Computing Machinery (ACM) Digital Library, and Institute of Electrical and Electronics Engineers (IEEE) Xplore Digital Library. After screening 362 articles, 48 papers were included for a comprehensive data collection. We noticed a typical workflow starting with feeding raw data, applying deep learning models, and ending with clinical outcome predictions as evaluations of the learned representations. Specifically, learning representations from structured EHR data was dominant (36 out of 48 studies). Recurrent Neural Networks were widely applied as the deep learning architecture (LSTM: 13 studies, GRU: 11 studies). Disease prediction was the most common application and evaluation (30 studies). Benchmark datasets were mostly unavailable (28 studies) due to privacy concerns of EHR data, and code availability was assured in 20 studies. We show the importance and feasibility of learning comprehensive representations of patient EHR data through a systematic review. Advances in patient representation learning techniques will be essential for powering patient-level EHR analyses. Future work will still be devoted to leveraging the richness and potential of available EHR data. Knowledge distillation and advanced learning techniques will be exploited to assist the capability of learning patient representation further.
Extracting Concepts for Precision Oncology from the Biomedical Literature
Sep 30, 2020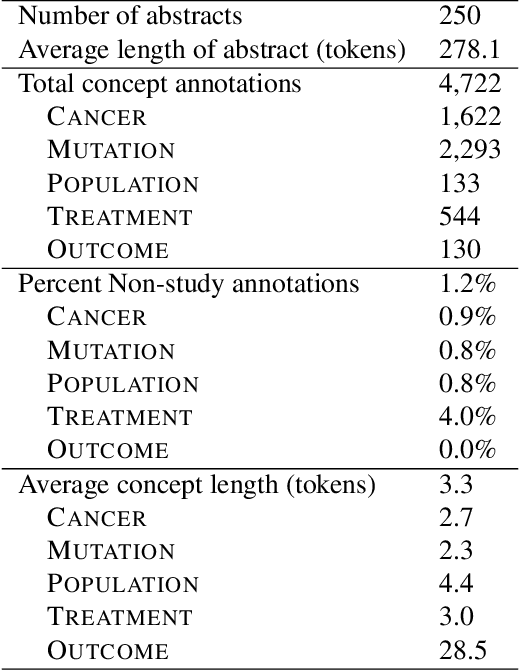
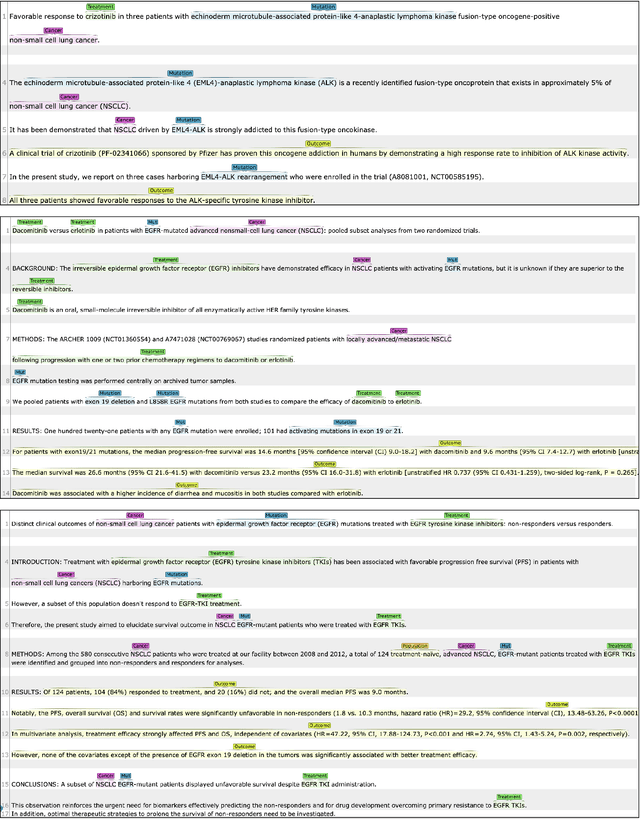

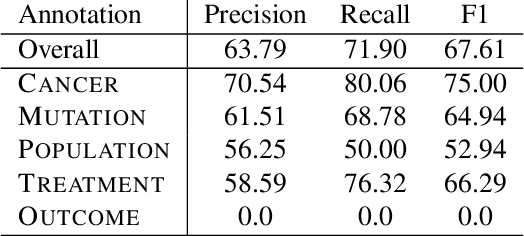
Abstract:This paper describes an initial dataset and automatic natural language processing (NLP) method for extracting concepts related to precision oncology from biomedical research articles. We extract five concept types: Cancer, Mutation, Population, Treatment, Outcome. A corpus of 250 biomedical abstracts were annotated with these concepts following standard double-annotation procedures. We then experiment with BERT-based models for concept extraction. The best-performing model achieved a precision of 63.8%, a recall of 71.9%, and an F1 of 67.1. Finally, we propose additional directions for research for improving extraction performance and utilizing the NLP system in downstream precision oncology applications.
Understanding Spatial Language in Radiology: Representation Framework, Annotation, and Spatial Relation Extraction from Chest X-ray Reports using Deep Learning
Aug 13, 2019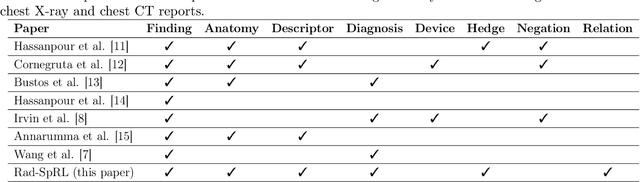
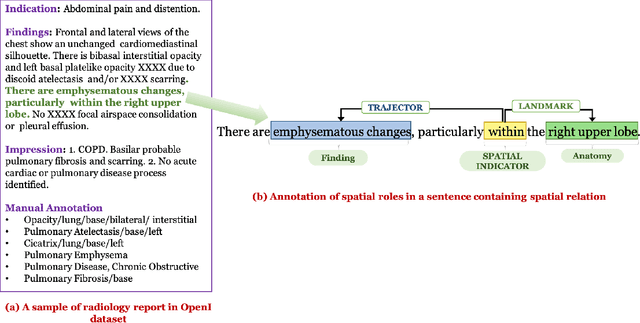

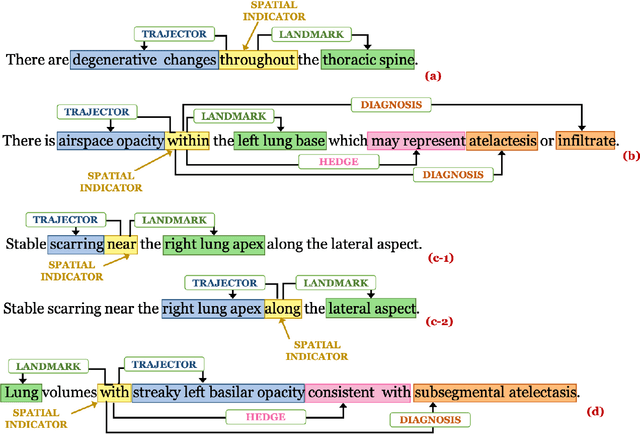
Abstract:We define a representation framework for extracting spatial information from radiology reports (Rad-SpRL). We annotated a total of 2000 chest X-ray reports with 4 spatial roles corresponding to the common radiology entities. Our focus is on extracting detailed information of a radiologist's interpretation containing a radiographic finding, its anatomical location, corresponding probable diagnoses, as well as associated hedging terms. For this, we propose a deep learning-based natural language processing (NLP) method involving both word and character-level encodings. Specifically, we utilize a bidirectional long short-term memory (Bi-LSTM) conditional random field (CRF) model for extracting the spatial roles. The model achieved average F1 measures of 90.28 and 94.61 for extracting the Trajector and Landmark roles respectively whereas the performance was moderate for Diagnosis and Hedge roles with average F1 of 71.47 and 73.27 respectively. The corpus will soon be made available upon request.
Enhancing Clinical Concept Extraction with Contextual Embedding
Feb 22, 2019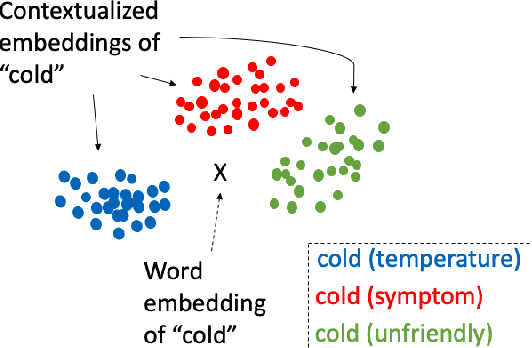



Abstract:Neural network-based representations ("embeddings") have dramatically advanced natural language processing (NLP) tasks in the past few years. This certainly holds for clinical concept extraction, especially when combined with deep learning-based models. Recently, however, more advanced embedding methods and representations (e.g., ELMo, BERT) have further pushed the state-of-the-art in NLP. While these certainly improve clinical concept extraction as well, there are no commonly agreed upon best practices for how to integrate these representations for extracting concepts. The purpose of this study, then, is to explore the space of possible options in utilizing these new models, including comparing these to more traditional word embedding methods (word2vec, GloVe, fastText). We evaluate a battery of embedding methods on four clinical concept extraction corpora, explore effects of pre-training on extraction performance, and present an intuitive way to understand the semantic information encoded by advanced contextualized representations. Notably, we achieved new state-of-the-art performances across all four corpora.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge