Surabhi Datta
Eye-SpatialNet: Spatial Information Extraction from Ophthalmology Notes
May 19, 2023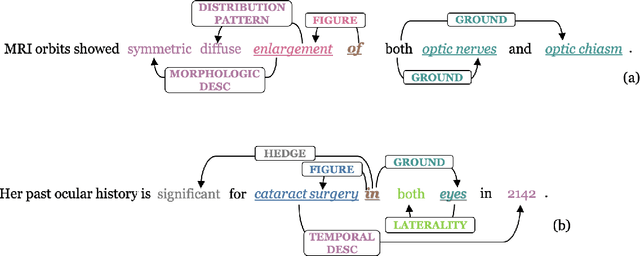



Abstract:We introduce an annotated corpus of 600 ophthalmology notes labeled with detailed spatial and contextual information of ophthalmic entities. We extend our previously proposed frame semantics-based spatial representation schema, Rad-SpatialNet, to represent spatial language in ophthalmology text, resulting in the Eye-SpatialNet schema. The spatially-grounded entities are findings, procedures, and drugs. To accurately capture all spatial details, we add some domain-specific elements in Eye-SpatialNet. The annotated corpus contains 1715 spatial triggers, 7308 findings, 2424 anatomies, and 9914 descriptors. To automatically extract the spatial information, we employ a two-turn question answering approach based on the transformer language model BERT. The results are promising, with F1 scores of 89.31, 74.86, and 88.47 for spatial triggers, Figure, and Ground frame elements, respectively. This is the first work to represent and extract a wide variety of clinical information in ophthalmology. Extracting detailed information can benefit ophthalmology applications and research targeted toward disease progression and screening.
Leveraging Spatial Information in Radiology Reports for Ischemic Stroke Phenotyping
Oct 10, 2020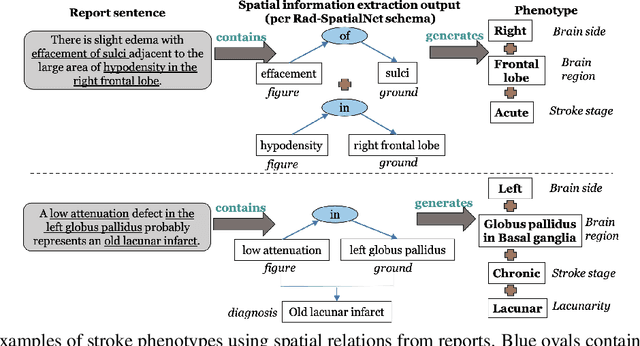
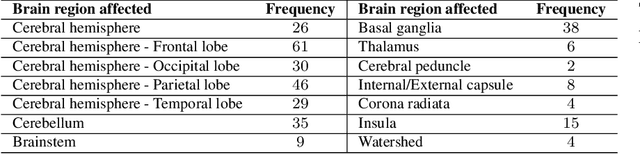
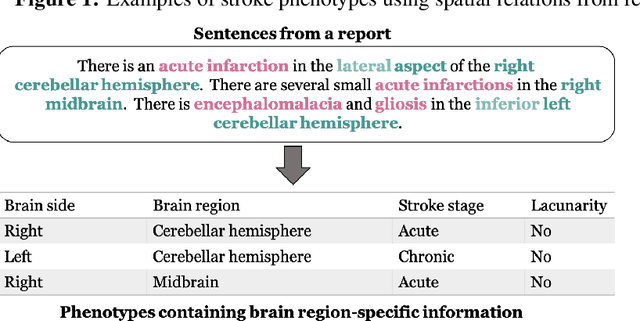

Abstract:Classifying fine-grained ischemic stroke phenotypes relies on identifying important clinical information. Radiology reports provide relevant information with context to determine such phenotype information. We focus on stroke phenotypes with location-specific information: brain region affected, laterality, stroke stage, and lacunarity. We use an existing fine-grained spatial information extraction system--Rad-SpatialNet--to identify clinically important information and apply simple domain rules on the extracted information to classify phenotypes. The performance of our proposed approach is promising (recall of 89.62% for classifying brain region and 74.11% for classifying brain region, side, and stroke stage together). Our work demonstrates that an information extraction system based on a fine-grained schema can be utilized to determine complex phenotypes with the inclusion of simple domain rules. These phenotypes have the potential to facilitate stroke research focusing on post-stroke outcome and treatment planning based on the stroke location.
RadLex Normalization in Radiology Reports
Sep 10, 2020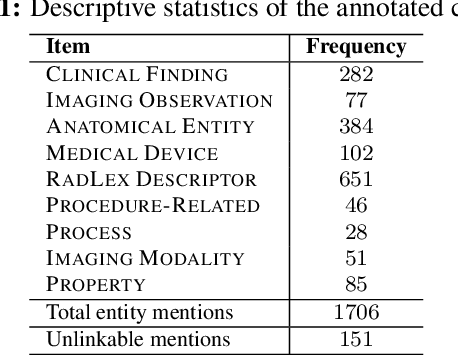
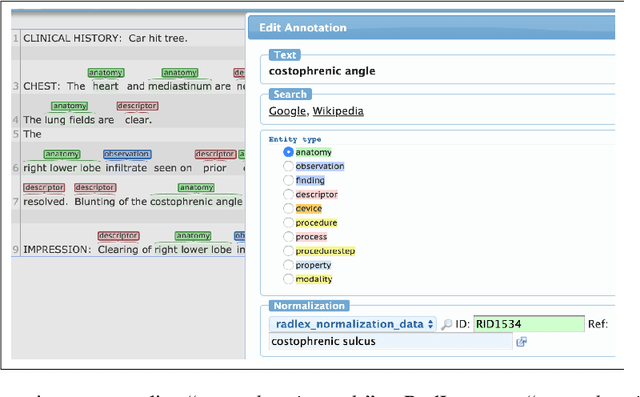
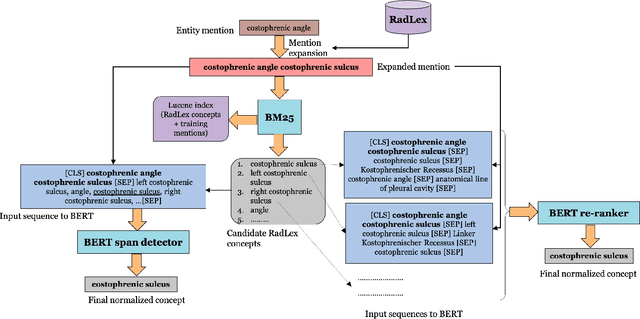

Abstract:Radiology reports have been widely used for extraction of various clinically significant information about patients' imaging studies. However, limited research has focused on standardizing the entities to a common radiology-specific vocabulary. Further, no study to date has attempted to leverage RadLex for standardization. In this paper, we aim to normalize a diverse set of radiological entities to RadLex terms. We manually construct a normalization corpus by annotating entities from three types of reports. This contains 1706 entity mentions. We propose two deep learning-based NLP methods based on a pre-trained language model (BERT) for automatic normalization. First, we employ BM25 to retrieve candidate concepts for the BERT-based models (re-ranker and span detector) to predict the normalized concept. The results are promising, with the best accuracy (78.44%) obtained by the span detector. Additionally, we discuss the challenges involved in corpus construction and propose new RadLex terms.
Understanding Spatial Language in Radiology: Representation Framework, Annotation, and Spatial Relation Extraction from Chest X-ray Reports using Deep Learning
Aug 13, 2019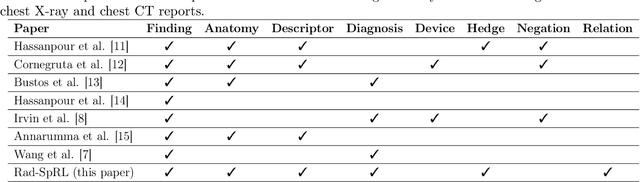
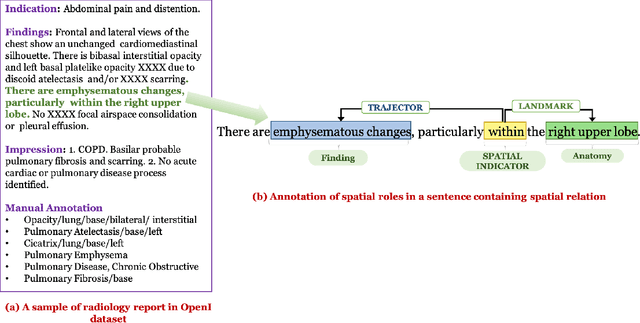

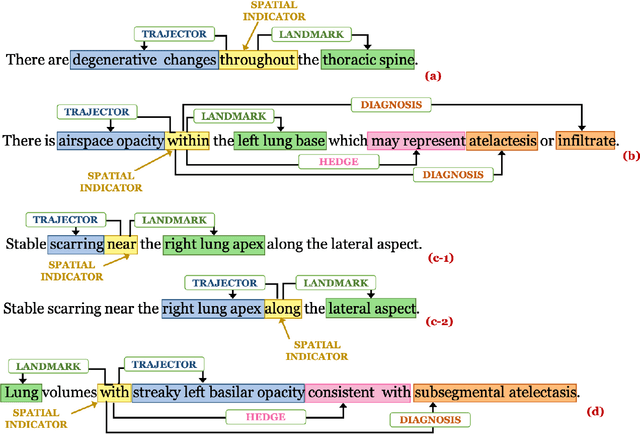
Abstract:We define a representation framework for extracting spatial information from radiology reports (Rad-SpRL). We annotated a total of 2000 chest X-ray reports with 4 spatial roles corresponding to the common radiology entities. Our focus is on extracting detailed information of a radiologist's interpretation containing a radiographic finding, its anatomical location, corresponding probable diagnoses, as well as associated hedging terms. For this, we propose a deep learning-based natural language processing (NLP) method involving both word and character-level encodings. Specifically, we utilize a bidirectional long short-term memory (Bi-LSTM) conditional random field (CRF) model for extracting the spatial roles. The model achieved average F1 measures of 90.28 and 94.61 for extracting the Trajector and Landmark roles respectively whereas the performance was moderate for Diagnosis and Hedge roles with average F1 of 71.47 and 73.27 respectively. The corpus will soon be made available upon request.
A frame semantic overview of NLP-based information extraction for cancer-related EHR notes
Apr 02, 2019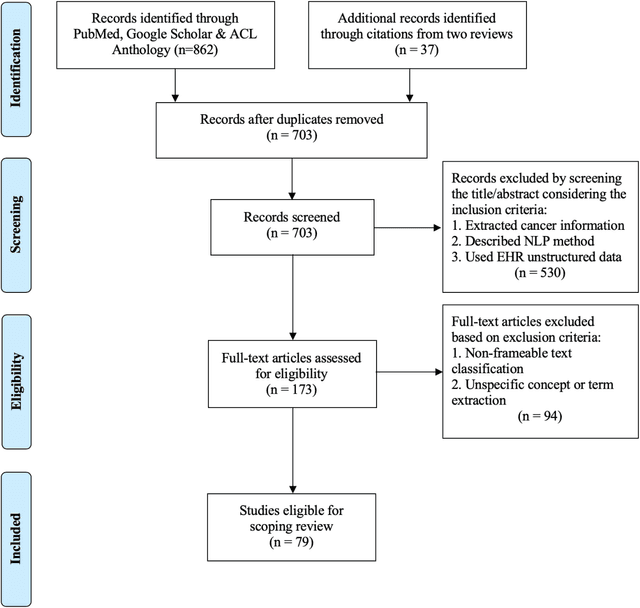
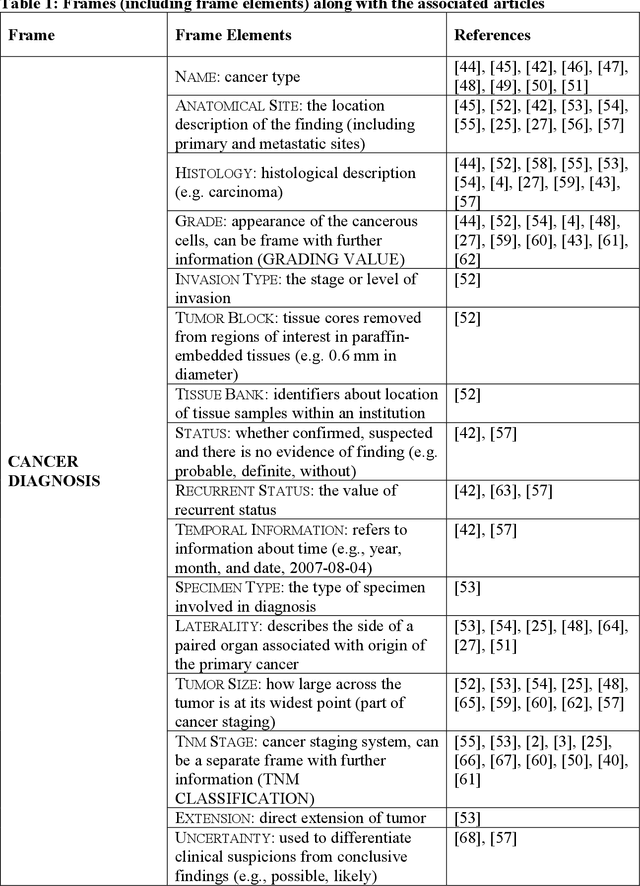
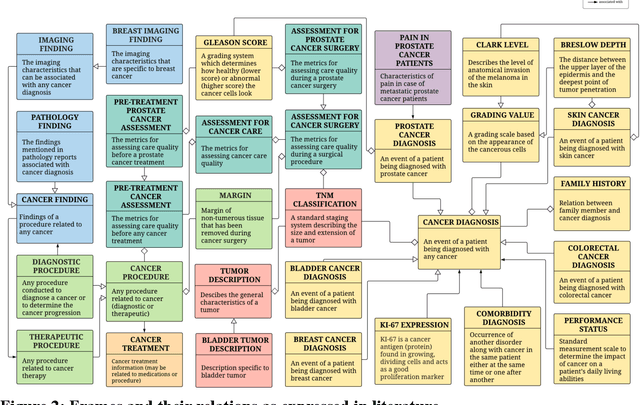
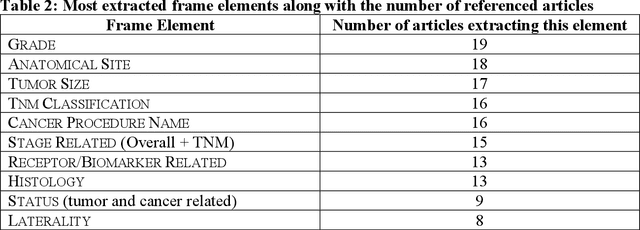
Abstract:Objective: There is a lot of information about cancer in Electronic Health Record (EHR) notes that can be useful for biomedical research provided natural language processing (NLP) methods are available to extract and structure this information. In this paper, we present a scoping review of existing clinical NLP literature for cancer. Methods: We identified studies describing an NLP method to extract specific cancer-related information from EHR sources from PubMed, Google Scholar, ACL Anthology, and existing reviews. Two exclusion criteria were used in this study. We excluded articles where the extraction techniques used were too broad to be represented as frames and also where very low-level extraction methods were used. 79 articles were included in the final review. We organized this information according to frame semantic principles to help identify common areas of overlap and potential gaps. Results: Frames were created from the reviewed articles pertaining to cancer information such as cancer diagnosis, tumor description, cancer procedure, breast cancer diagnosis, prostate cancer diagnosis and pain in prostate cancer patients. These frames included both a definition as well as specific frame elements (i.e. extractable attributes). We found that cancer diagnosis was the most common frame among the reviewed papers (36 out of 79), with recent work focusing on extracting information related to treatment and breast cancer diagnosis. Conclusion: The list of common frames described in this paper identifies important cancer-related information extracted by existing NLP techniques and serves as a useful resource for future researchers requiring cancer information extracted from EHR notes. We also argue, due to the heavy duplication of cancer NLP systems, that a general purpose resource of annotated cancer frames and corresponding NLP tools would be valuable.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge