Kirk Roberts
Gender-Neutral Large Language Models for Medical Applications: Reducing Bias in PubMed Abstracts
Jan 10, 2025



Abstract:This paper presents a pipeline for mitigating gender bias in large language models (LLMs) used in medical literature by neutralizing gendered occupational pronouns. A dataset of 379,000 PubMed abstracts from 1965-1980 was processed to identify and modify pronouns tied to professions. We developed a BERT-based model, ``Modern Occupational Bias Elimination with Refined Training,'' or ``MOBERT,'' trained on these neutralized abstracts, and compared its performance with ``1965Bert,'' trained on the original dataset. MOBERT achieved a 70\% inclusive replacement rate, while 1965Bert reached only 4\%. A further analysis of MOBERT revealed that pronoun replacement accuracy correlated with the frequency of occupational terms in the training data. We propose expanding the dataset and refining the pipeline to improve performance and ensure more equitable language modeling in medical applications.
Overview of TREC 2024 Biomedical Generative Retrieval (BioGen) Track
Nov 27, 2024



Abstract:With the advancement of large language models (LLMs), the biomedical domain has seen significant progress and improvement in multiple tasks such as biomedical question answering, lay language summarization of the biomedical literature, clinical note summarization, etc. However, hallucinations or confabulations remain one of the key challenges when using LLMs in the biomedical and other domains. Inaccuracies may be particularly harmful in high-risk situations, such as making clinical decisions or appraising biomedical research. Studies on the evaluation of the LLMs' abilities to ground generated statements in verifiable sources have shown that models perform significantly worse on lay-user generated questions, and often fail to reference relevant sources. This can be problematic when those seeking information want evidence from studies to back up the claims from LLMs[3]. Unsupported statements are a major barrier to using LLMs in any applications that may affect health. Methods for grounding generated statements in reliable sources along with practical evaluation approaches are needed to overcome this barrier. Towards this, in our pilot task organized at TREC 2024, we introduced the task of reference attribution as a means to mitigate the generation of false statements by LLMs answering biomedical questions.
LLM-IE: A Python Package for Generative Information Extraction with Large Language Models
Nov 18, 2024



Abstract:Objectives: Despite the recent adoption of large language models (LLMs) for biomedical information extraction, challenges in prompt engineering and algorithms persist, with no dedicated software available. To address this, we developed LLM-IE: a Python package for building complete information extraction pipelines. Our key innovation is an interactive LLM agent to support schema definition and prompt design. Materials and Methods: The LLM-IE supports named entity recognition, entity attribute extraction, and relation extraction tasks. We benchmarked on the i2b2 datasets and conducted a system evaluation. Results: The sentence-based prompting algorithm resulted in the best performance while requiring a longer inference time. System evaluation provided intuitive visualization. Discussion: LLM-IE was designed from practical NLP experience in healthcare and has been adopted in internal projects. It should hold great value to the biomedical NLP community. Conclusion: We developed a Python package, LLM-IE, that provides building blocks for robust information extraction pipeline construction.
A Comparative Study of Recent Large Language Models on Generating Hospital Discharge Summaries for Lung Cancer Patients
Nov 06, 2024


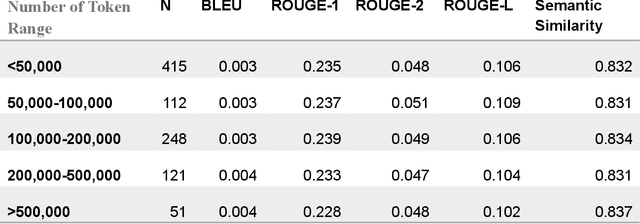
Abstract:Generating discharge summaries is a crucial yet time-consuming task in clinical practice, essential for conveying pertinent patient information and facilitating continuity of care. Recent advancements in large language models (LLMs) have significantly enhanced their capability in understanding and summarizing complex medical texts. This research aims to explore how LLMs can alleviate the burden of manual summarization, streamline workflow efficiencies, and support informed decision-making in healthcare settings. Clinical notes from a cohort of 1,099 lung cancer patients were utilized, with a subset of 50 patients for testing purposes, and 102 patients used for model fine-tuning. This study evaluates the performance of multiple LLMs, including GPT-3.5, GPT-4, GPT-4o, and LLaMA 3 8b, in generating discharge summaries. Evaluation metrics included token-level analysis (BLEU, ROUGE-1, ROUGE-2, ROUGE-L) and semantic similarity scores between model-generated summaries and physician-written gold standards. LLaMA 3 8b was further tested on clinical notes of varying lengths to examine the stability of its performance. The study found notable variations in summarization capabilities among LLMs. GPT-4o and fine-tuned LLaMA 3 demonstrated superior token-level evaluation metrics, while LLaMA 3 consistently produced concise summaries across different input lengths. Semantic similarity scores indicated GPT-4o and LLaMA 3 as leading models in capturing clinical relevance. This study contributes insights into the efficacy of LLMs for generating discharge summaries, highlighting LLaMA 3's robust performance in maintaining clarity and relevance across varying clinical contexts. These findings underscore the potential of automated summarization tools to enhance documentation precision and efficiency, ultimately improving patient care and operational capability in healthcare settings.
Exploring the Generalization of Cancer Clinical Trial Eligibility Classifiers Across Diseases
Mar 25, 2024


Abstract:Clinical trials are pivotal in medical research, and NLP can enhance their success, with application in recruitment. This study aims to evaluate the generalizability of eligibility classification across a broad spectrum of clinical trials. Starting with phase 3 cancer trials, annotated with seven eligibility exclusions, then to determine how well models can generalize to non-cancer and non-phase 3 trials. To assess this, we have compiled eligibility criteria data for five types of trials: (1) additional phase 3 cancer trials, (2) phase 1 and 2 cancer trials, (3) heart disease trials, (4) type 2 diabetes trials, and (5) observational trials for any disease, comprising 2,490 annotated eligibility criteria across seven exclusion types. Our results show that models trained on the extensive cancer dataset can effectively handle criteria commonly found in non-cancer trials, such as autoimmune diseases. However, they struggle with criteria disproportionately prevalent in cancer trials, like prior malignancy. We also experiment with few-shot learning, demonstrating that a limited number of disease-specific examples can partially overcome this performance gap. We are releasing this new dataset of annotated eligibility statements to promote the development of cross-disease generalization in clinical trial classification.
Question Answering for Electronic Health Records: A Scoping Review of datasets and models
Oct 12, 2023



Abstract:Question Answering (QA) systems on patient-related data can assist both clinicians and patients. They can, for example, assist clinicians in decision-making and enable patients to have a better understanding of their medical history. Significant amounts of patient data are stored in Electronic Health Records (EHRs), making EHR QA an important research area. In EHR QA, the answer is obtained from the medical record of the patient. Because of the differences in data format and modality, this differs greatly from other medical QA tasks that employ medical websites or scientific papers to retrieve answers, making it critical to research EHR question answering. This study aimed to provide a methodological review of existing works on QA over EHRs. We searched for articles from January 1st, 2005 to September 30th, 2023 in four digital sources including Google Scholar, ACL Anthology, ACM Digital Library, and PubMed to collect relevant publications on EHR QA. 4111 papers were identified for our study, and after screening based on our inclusion criteria, we obtained a total of 47 papers for further study. Out of the 47 papers, 25 papers were about EHR QA datasets, and 37 papers were about EHR QA models. It was observed that QA on EHRs is relatively new and unexplored. Most of the works are fairly recent. Also, it was observed that emrQA is by far the most popular EHR QA dataset, both in terms of citations and usage in other papers. Furthermore, we identified the different models used in EHR QA along with the evaluation metrics used for these models.
Text Classification of Cancer Clinical Trial Eligibility Criteria
Sep 15, 2023Abstract:Automatic identification of clinical trials for which a patient is eligible is complicated by the fact that trial eligibility is stated in natural language. A potential solution to this problem is to employ text classification methods for common types of eligibility criteria. In this study, we focus on seven common exclusion criteria in cancer trials: prior malignancy, human immunodeficiency virus, hepatitis B, hepatitis C, psychiatric illness, drug/substance abuse, and autoimmune illness. Our dataset consists of 764 phase III cancer trials with these exclusions annotated at the trial level. We experiment with common transformer models as well as a new pre-trained clinical trial BERT model. Our results demonstrate the feasibility of automatically classifying common exclusion criteria. Additionally, we demonstrate the value of a pre-trained language model specifically for clinical trials, which yields the highest average performance across all criteria.
Evaluation of AI Chatbots for Patient-Specific EHR Questions
Jun 05, 2023



Abstract:This paper investigates the use of artificial intelligence chatbots for patient-specific question answering (QA) from clinical notes using several large language model (LLM) based systems: ChatGPT (versions 3.5 and 4), Google Bard, and Claude. We evaluate the accuracy, relevance, comprehensiveness, and coherence of the answers generated by each model using a 5-point Likert scale on a set of patient-specific questions.
Eye-SpatialNet: Spatial Information Extraction from Ophthalmology Notes
May 19, 2023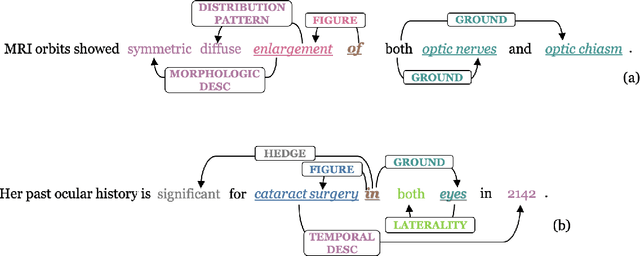



Abstract:We introduce an annotated corpus of 600 ophthalmology notes labeled with detailed spatial and contextual information of ophthalmic entities. We extend our previously proposed frame semantics-based spatial representation schema, Rad-SpatialNet, to represent spatial language in ophthalmology text, resulting in the Eye-SpatialNet schema. The spatially-grounded entities are findings, procedures, and drugs. To accurately capture all spatial details, we add some domain-specific elements in Eye-SpatialNet. The annotated corpus contains 1715 spatial triggers, 7308 findings, 2424 anatomies, and 9914 descriptors. To automatically extract the spatial information, we employ a two-turn question answering approach based on the transformer language model BERT. The results are promising, with F1 scores of 89.31, 74.86, and 88.47 for spatial triggers, Figure, and Ground frame elements, respectively. This is the first work to represent and extract a wide variety of clinical information in ophthalmology. Extracting detailed information can benefit ophthalmology applications and research targeted toward disease progression and screening.
Application of an ontology for model cards to generate computable artifacts for linking machine learning information from biomedical research
Mar 21, 2023



Abstract:Model card reports provide a transparent description of machine learning models which includes information about their evaluation, limitations, intended use, etc. Federal health agencies have expressed an interest in model cards report for research studies using machine-learning based AI. Previously, we have developed an ontology model for model card reports to structure and formalize these reports. In this paper, we demonstrate a Java-based library (OWL API, FaCT++) that leverages our ontology to publish computable model card reports. We discuss future directions and other use cases that highlight applicability and feasibility of ontology-driven systems to support FAIR challenges.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge